-
PDF
- Split View
-
Views
-
Cite
Cite
Jonathan M. Guberman, J. Ai, O. Arnaiz, Joachim Baran, Andrew Blake, Richard Baldock, Claude Chelala, David Croft, Anthony Cros, Rosalind J. Cutts, A. Di Génova, Simon Forbes, T. Fujisawa, E. Gadaleta, D. M. Goodstein, Gunes Gundem, Bernard Haggarty, Syed Haider, Matthew Hall, Todd Harris, Robin Haw, S. Hu, Simon Hubbard, Jack Hsu, Vivek Iyer, Philip Jones, Toshiaki Katayama, R. Kinsella, Lei Kong, Daniel Lawson, Yong Liang, Nuria Lopez-Bigas, J. Luo, Michael Lush, Jeremy Mason, Francois Moreews, Nelson Ndegwa, Darren Oakley, Christian Perez-Llamas, Michael Primig, Elena Rivkin, S. Rosanoff, Rebecca Shepherd, Reinhard Simon, B. Skarnes, Damian Smedley, Linda Sperling, William Spooner, Peter Stevenson, Kevin Stone, J. Teague, Jun Wang, Jianxin Wang, Brett Whitty, D. T. Wong, Marie Wong-Erasmus, L. Yao, Ken Youens-Clark, Christina Yung, Junjun Zhang, Arek Kasprzyk, BioMart Central Portal: an open database network for the biological community, Database, Volume 2011, 2011, bar041, https://doi.org/10.1093/database/bar041
Close - Share Icon Share
Abstract
BioMart Central Portal is a first of its kind, community-driven effort to provide unified access to dozens of biological databases spanning genomics, proteomics, model organisms, cancer data, ontology information and more. Anybody can contribute an independently maintained resource to the Central Portal, allowing it to be exposed to and shared with the research community, and linking it with the other resources in the portal. Users can take advantage of the common interface to quickly utilize different sources without learning a new system for each. The system also simplifies cross-database searches that might otherwise require several complicated steps. Several integrated tools streamline common tasks, such as converting between ID formats and retrieving sequences. The combination of a wide variety of databases, an easy-to-use interface, robust programmatic access and the array of tools make Central Portal a one-stop shop for biological data querying. Here, we describe the structure of Central Portal and show example queries to demonstrate its capabilities.
Database URL: http://central.biomart.org.
Project description
Introduction
BioMart is a free, open-source, federated database system (1–3). It is cross-platform and supports many popular relational database managements systems, including MySQL, Oracle, PostgreSQL, SQL Server and DB2. The software is data-agnostic, and can therefore be easily adapted to existing data sets. It is expandable and customizable through a plug-in system, and is open-source so the community can participate in deeper development. Furthermore, BioMart can seamlessly connect geographically disparate databases, facilitating collaboration between different groups. These features have catalyzed the creation of BioMart Central Portal, a first of its kind community-supported effort to create a single access point integrating many different, independently administered biological databases (Figure 1).
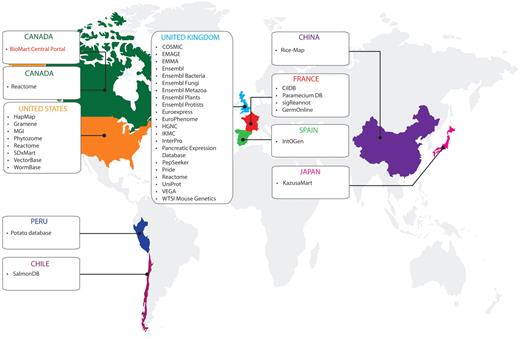
Databases available on the BioMart Central Portal and their host countries (April 2011).
For administrators, participation in Central Portal offers several benefits. Central Portal can provide an instantly available and automatically updated source of annotations for other projects, as is done in the International Cancer Genome Consortium Data Portal (4). Being part of the community can also expose a database to a wide user base. Furthermore, because the BioMart software allows administrators to easily create their own plug-ins, joining the community allows administrators to take advantage of the tools that others have created, thereby enhancing their own databases. Central Portal passes queries directly to the individual member servers, so administrators retain full control of their databases and their data (Figure 2).
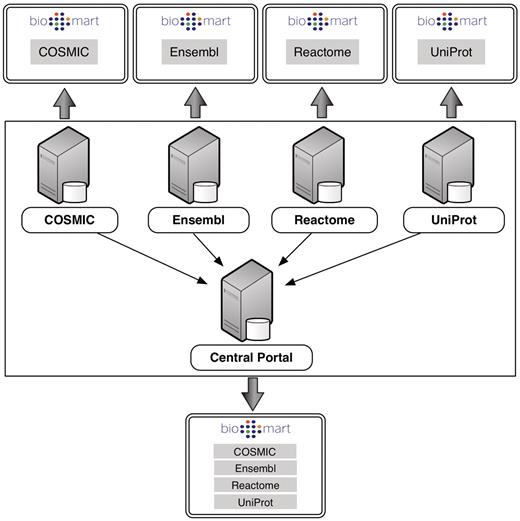
Each individual server hosts its own instance of BioMart retrieving data from its own local database backend. Central Portal offers a unified access point to all of these databases, distributing queries to the appropriate servers.
For users, Central Portal offers a central repository for a vast array of biological data. BioMart can interoperate with other web sites, because results can be configured to link to outside resources; examples in Central Portal include KEGG pathway information (5–7) and Pancreatic Expression Database entries (8). The intuitive interface is consistent across all databases, so users familiar with one source can immediately transfer their skills to another data source. Since Central Portal is constantly updated, users are immediately exposed to new resources as they become available. In addition to the web-based interface, Central Portal also offers a wide variety of other access methods for more advanced querying, including application programming interfaces (APIs) for Java, SPARQL, REST and SOAP.
Moreover, both users and administrators benefit from the value gained by having individual databases connected in a central access point. By allowing data sets to be linked together, resources can be combined in novel ways, potentially revealing unexpected connections or suggesting new avenues of inquiry. The strength of the Central Portal comes from the fact that it is created and supported by a large community, and, as a whole, it is greater than the sum of its parts.
Interface
When viewing the Central Portal home page, users are presented with the main querying section, which is divided into three subsections: Identifier Search, Tools and Database Search (Figure 3).
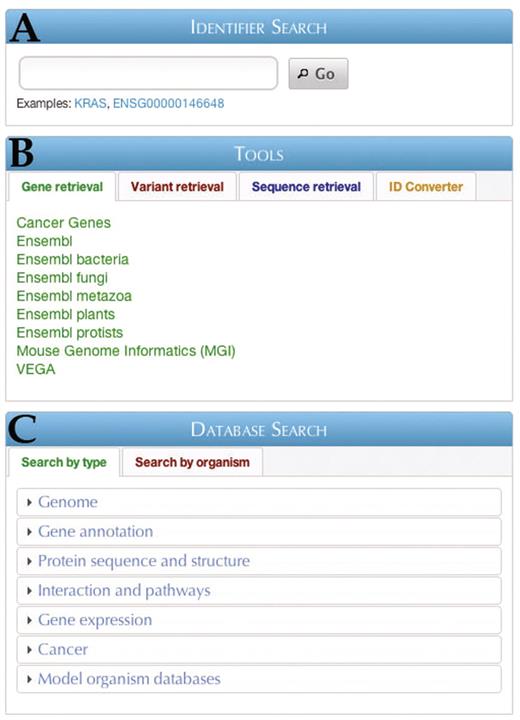
The BioMart Central Portal home page. Three main entry points are available: (A) Identifier search, (B) Tools and (C) Database search.
The Identifier Search (Figure 3A) allows users to input gene identifiers in a number of formats (e.g. Gene name, Ensembl IDs, RefSeq IDs, etc.) and search for it across all of the member databases in the Portal. The result of the search links to a report page for the identifier, which summarizes key information about the search term taken from several sources (Figure 4). With this function users can quickly find information about a single identifier, and perhaps even locate resources that they did not realize were applicable to the target of their query.
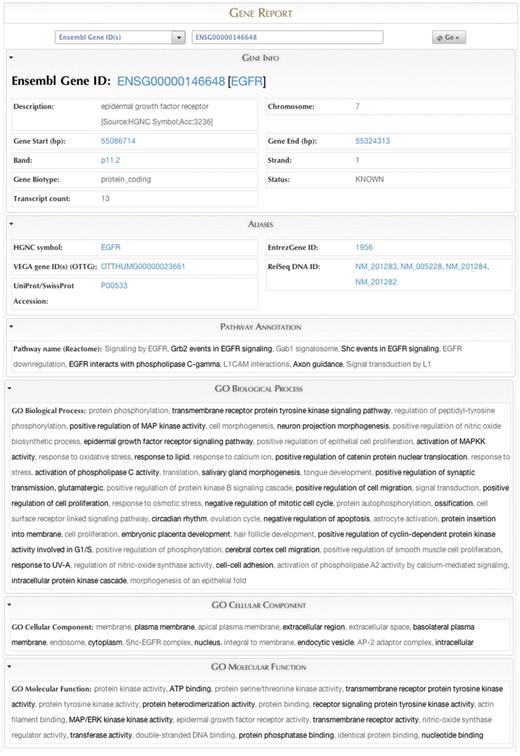
The Gene Report page for EGFR, displaying data federated from several sources.
The Tools section (Figure 3B) contains links to various data analysis tools in four categories: Gene retrieval, Variant retrieval, Sequence retrieval and ID Converter. The first two sections allow quick access to some of the largest and most popular databases contained in Central Portal. The third section, Sequence retrieval, allows easy querying of genomic and protein sequences in any of several formats (Figure 5). The fourth section, the ID Converter tool, allows users to enter or upload a list of identifiers in any format supported by a BioMart database, and retrieve the same list converted to any other supported format.
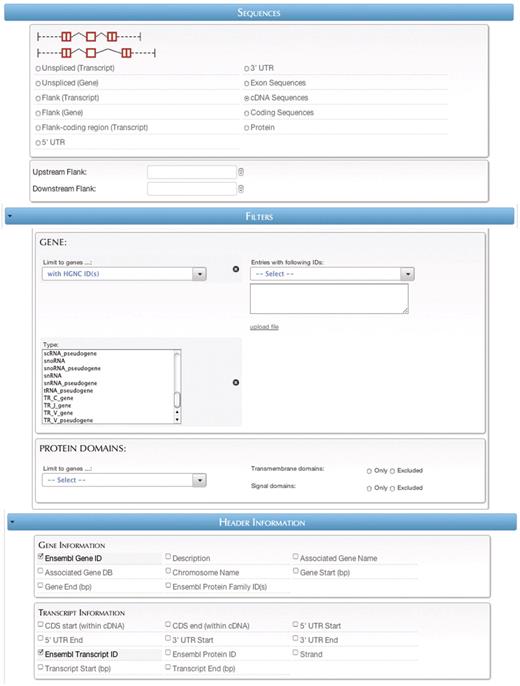
In the Database Search section (Figure 3C), users can access the individual member databases for querying through the BioMart interface. To make finding the relevant database easier, users can choose to browse databases by the type of information contained therein (Search by type) or by the organism with which the database is concerned (Search by organism). Browse by type is further subdivided into several categories such as Genome [e.g. Ensembl databases (9)], Gene annotation [e.g. HGNC (10)], Protein sequence and structure [e.g. InterPro (11)], Interactions and pathways [e.g. Reactome (12)], Gene expression [e.g. EMAGE (13)], Cancer [e.g. COSMIC (14)] and Model organism databases [e.g. Gramene (15)], Search by organism is subdivided into categories for bacteria, plants, protists, invertebrates and vertebrates. After choosing a data set, users can construct queries using the basic BioMart concepts of attributes, which indicate what information should be returned, and filters, which restrict the database entries that are retrieved.
Access methods
In addition to the graphical user interfaces, Central Portal also offers programmatic access to allow for automated querying. Several programming interfaces are available: an XML querying method that can be accessed via REST or SOAP requests, a full Java API and RDF querying via SPARQL. The syntax of any of the APIs is easy to use for programmers familiar with the basic BioMart concepts of attributes, filters and data sets. For example, to retrieve a list of filters for a given data set, a client could use the REST API and access the URL /martservice/filters?datasets=datasetname. Alternatively and equivalently, the client could use the Java API using the method getFilters(datasetname) to accomplish the same result. Because, there are a variety of APIs available, developers can choose the access method that makes the most sense for their specific applications and use cases.
To further ease the adoption of the APIs, the equivalent code of any query constructed in the web GUI can be retrieved in any of the API formats by clicking on the appropriate button on the query page; in this way, queries can be saved, modified and easily transferred from one format to another. It also provides a readily available graphical method of constructing complex API calls, which could be of use in certain tools or scripts.
Data content
BioMart Central Portal contains a constantly growing list of data sources accessible by a wide variety of methods and tools. The following table reflects the contents of the portal as of May 2011:
| Database . | Location . | Description . | References . |
|---|---|---|---|
| Cildb | CNRS, France | Database for eukaryotic cilia and centriolar structures, integrating orthology relationships for 33 species with high-throughput studies and OMIM | (16) |
| COSMIC | WTSI, UK | Somatic mutation information relating to human cancers | (14) |
| EMAGE | MRC HGU, UK | In situ gene expression data in the mouse embryo | (13) |
| EMMA | EBI, UK | Mouse mutant strain information | (17) |
| Ensembl | WTSI/EBI, UK | Genome databases for vertebrates and other eukaryotic species | (9) |
| Ensembl Bacteria | EBI, UK | Genome databases for bacteria | (9) |
| Ensembl Fungi | EBI, UK | Genome databases for fungi | (9) |
| Ensembl Metazoa | EBI, UK | Genome databases for metazoa | (9) |
| Ensembl Plants | EBI, UK | Genome databases for plants | (9) |
| Ensembl Protists | EBI, UK | Genome databases for protists | (9) |
| Eurexpress | MRC HGU, UK | Transcriptome atlas database for mouse embryo | (18) |
| EuroPhenome | MRC Harwell, UK | Mouse phenotyping data | (19) |
| GermOnline | Inserm, France | Cross-species microarray expression database focusing on germline development, meiosis and gametogenesis as well as the mitotic cell cycle | (20) |
| Gramene | CSHL, USA | Agriculturally important grass genomes | (15) |
| HapMap | NCBI, USA | Multi-country effort to identify and catalog genetic similarities and differences in human beings | (21) |
| HGNC | EBI, UK | Repository of human gene nomenclature and associated resources | (10) |
| IKMC | WTSI, UK | Data on mutant products (mice, ES cells and vectors) generated and made available by members of the International Knockout Mouse Consotium | (22) |
| InterPro | EBI, UK | Integrated database of predictive protein ‘signatures’ used for the classification and automatic annotation of proteins and genomes | (11) |
| IntOGen | UPF, Spain | Integrated multi-dimensional data for the identification of genes and groups of genes involved in cancer development | (23) |
| KazusaMart | Kazusa, Japan | Cyanobase, rhizobia and plant genome databases | (24) |
| MGI | Jackson Laboratory, USA | Mouse genome features, locations, alleles and orthologues | (25) |
| Pancreatic Expression Database | Barts Cancer Institute, UK | Results from published pancreatic cancer papers | (8) |
| Paramecium DB | CNRS, France | Paramecium genome database | (26) |
| PepSeeker | University of Manchester, UK | Database of proteome peptide identifications for investigating fragmentation patterns | (27) |
| Phytozome | JGI/CIG, USA | Comparative genomics of green plants | (28) |
| Potato Database | CIP, Peru | Potato and sweet potato phenotypic and genomic information | (29) |
| PRIDE | EBI, UK | Repository for protein and peptide identifications | (30) |
| Reactome | OICR, Canada; EBI, UK; NYU Medical Center, USA | Curated pathway annotation database | (12) |
| Rice-Map | Peking University, China | Rice (japonica and indica) genome annotation database | (31) |
| SalmonDB | CMM, Chile | Genomic information for Atlantic salmon, rainbow trout and related species | (32) |
| SDxMart | UCLA, USA | Saliva diagnostics for high-impact human diseases | (33) |
| sigReannot | Rennes, France | Aquaculture and farm animal species EST contigs | (34) |
| UniProt | EBI, UK | Protein sequence and functional information | (35) |
| VectorBase | University of Notre Dame, USA | Genome information for invertebrate vectors of human pathogens | (36) |
| VEGA | WTSI, UK | Manual annotation of vertebrate genome sequences | (37) |
| WormBase | California Institute of Technology, USA; CSHL, USA; EBI, UK; Washington University, USA | Caenorhabditis elegans and related nematode genomic information | (38) |
| WTSI Mouse Genetics | WTSI, UK | Mouse phenotyping and expression data captured from mutant mouse lines | (39) |
| Database . | Location . | Description . | References . |
|---|---|---|---|
| Cildb | CNRS, France | Database for eukaryotic cilia and centriolar structures, integrating orthology relationships for 33 species with high-throughput studies and OMIM | (16) |
| COSMIC | WTSI, UK | Somatic mutation information relating to human cancers | (14) |
| EMAGE | MRC HGU, UK | In situ gene expression data in the mouse embryo | (13) |
| EMMA | EBI, UK | Mouse mutant strain information | (17) |
| Ensembl | WTSI/EBI, UK | Genome databases for vertebrates and other eukaryotic species | (9) |
| Ensembl Bacteria | EBI, UK | Genome databases for bacteria | (9) |
| Ensembl Fungi | EBI, UK | Genome databases for fungi | (9) |
| Ensembl Metazoa | EBI, UK | Genome databases for metazoa | (9) |
| Ensembl Plants | EBI, UK | Genome databases for plants | (9) |
| Ensembl Protists | EBI, UK | Genome databases for protists | (9) |
| Eurexpress | MRC HGU, UK | Transcriptome atlas database for mouse embryo | (18) |
| EuroPhenome | MRC Harwell, UK | Mouse phenotyping data | (19) |
| GermOnline | Inserm, France | Cross-species microarray expression database focusing on germline development, meiosis and gametogenesis as well as the mitotic cell cycle | (20) |
| Gramene | CSHL, USA | Agriculturally important grass genomes | (15) |
| HapMap | NCBI, USA | Multi-country effort to identify and catalog genetic similarities and differences in human beings | (21) |
| HGNC | EBI, UK | Repository of human gene nomenclature and associated resources | (10) |
| IKMC | WTSI, UK | Data on mutant products (mice, ES cells and vectors) generated and made available by members of the International Knockout Mouse Consotium | (22) |
| InterPro | EBI, UK | Integrated database of predictive protein ‘signatures’ used for the classification and automatic annotation of proteins and genomes | (11) |
| IntOGen | UPF, Spain | Integrated multi-dimensional data for the identification of genes and groups of genes involved in cancer development | (23) |
| KazusaMart | Kazusa, Japan | Cyanobase, rhizobia and plant genome databases | (24) |
| MGI | Jackson Laboratory, USA | Mouse genome features, locations, alleles and orthologues | (25) |
| Pancreatic Expression Database | Barts Cancer Institute, UK | Results from published pancreatic cancer papers | (8) |
| Paramecium DB | CNRS, France | Paramecium genome database | (26) |
| PepSeeker | University of Manchester, UK | Database of proteome peptide identifications for investigating fragmentation patterns | (27) |
| Phytozome | JGI/CIG, USA | Comparative genomics of green plants | (28) |
| Potato Database | CIP, Peru | Potato and sweet potato phenotypic and genomic information | (29) |
| PRIDE | EBI, UK | Repository for protein and peptide identifications | (30) |
| Reactome | OICR, Canada; EBI, UK; NYU Medical Center, USA | Curated pathway annotation database | (12) |
| Rice-Map | Peking University, China | Rice (japonica and indica) genome annotation database | (31) |
| SalmonDB | CMM, Chile | Genomic information for Atlantic salmon, rainbow trout and related species | (32) |
| SDxMart | UCLA, USA | Saliva diagnostics for high-impact human diseases | (33) |
| sigReannot | Rennes, France | Aquaculture and farm animal species EST contigs | (34) |
| UniProt | EBI, UK | Protein sequence and functional information | (35) |
| VectorBase | University of Notre Dame, USA | Genome information for invertebrate vectors of human pathogens | (36) |
| VEGA | WTSI, UK | Manual annotation of vertebrate genome sequences | (37) |
| WormBase | California Institute of Technology, USA; CSHL, USA; EBI, UK; Washington University, USA | Caenorhabditis elegans and related nematode genomic information | (38) |
| WTSI Mouse Genetics | WTSI, UK | Mouse phenotyping and expression data captured from mutant mouse lines | (39) |
Query examples
One of the great strengths of Central Portal is that it allows cross-database searches that any individual resource would not. Here are some examples of the possibilities afforded by this feature.
Query #1: ‘Find insertion-frameshift mutations in the COSMIC database that affect genes involved in Apoptosis’.
| Entry point . | Filters . |
|---|---|
| Gene retrieval > cancer genes | COSMIC: |
| Mutation type-AA: Insertion-frameshift | |
| KEGG: | |
| KEGG Pathway: apoptosis |
| Entry point . | Filters . |
|---|---|
| Gene retrieval > cancer genes | COSMIC: |
| Mutation type-AA: Insertion-frameshift | |
| KEGG: | |
| KEGG Pathway: apoptosis |
By integrating data from the COSMIC and KEGG databases, Central Portal allows users to identify COSMIC mutations specific to their pathways of interest. The Pathway title links back to the KEGG web site and mutation ID links back to the COSMIC web site, providing the ability to obtain more detailed information on the pathway or on the mutation, respectively.
Query #2: ‘Retrieve the cDNA sequences of protein-coding human genes that have HGNC IDs’ (Figure 5).
| Entry point . | Data sets . | Filters/attributes . |
|---|---|---|
| Sequence retrieval > Ensembl | Homo sapiens gene (GRCh37.p2) | Sequences: cDNA sequences |
| Filters: | ||
| Limit to genes: with HGNC ID(s) | ||
| Type: protein_coding | ||
| Header information: | ||
| Ensembl Gene ID | ||
| Ensembl Transcript ID |
| Entry point . | Data sets . | Filters/attributes . |
|---|---|---|
| Sequence retrieval > Ensembl | Homo sapiens gene (GRCh37.p2) | Sequences: cDNA sequences |
| Filters: | ||
| Limit to genes: with HGNC ID(s) | ||
| Type: protein_coding | ||
| Header information: | ||
| Ensembl Gene ID | ||
| Ensembl Transcript ID |
By combining the sequence retrieval tool with search capabilities, BioMart reduces what is often a two-step process—retrieving a list of genes, and then retrieving the sequences of those genes—into a single query.
Future directions
BioMart Central Portal is constantly evolving thanks to the efforts of the community that supports it and contributes data. To make joining Central Portal easier, we are creating BioMart Central Registry. With this resource, database administrators will be able to create an account, add their data sources and suggest categorization for them. Once registered, participants will also be able to make changes to their databases and notify Central Portal of updates.
In addition to including new data sets, Central Portal will evolve, as new tools are developed and added. Such tools will perform deeper analysis, such as detecting enrichment of certain properties (e.g. GO terms) within a given set of genes or calculating consequences given a list of SNP terms. BioMart plug-ins developed by other community members may also be incorporated, further strengthening the project as a whole.
Funding
The development of the BioMart software and the creation and hosting of BioMart Central Portal was supported by the Ontario Institute for Cancer Research and the Ontario Ministry for Research and Innovation. The individual data sources that Central Portal comprises are funded separately and independently.
Conflict of interest. None declared.
Acknowledgements
BioMart Central Portal is a collaborative, community effort and as such it is the product of the efforts of dozens, if not hundreds, of people. Creating a biological database is a multi-step process: experimenters must collect the data, database managers must create data models and administer databases and bioinformaticians must create methods for analysing the data. Additionally, over the years many programmers have contributed to the BioMart project codebase. We would like to acknowledge all the hard work of the many contributors to the projects that BioMart comprises.



