-
PDF
- Split View
-
Views
-
Cite
Cite
Zhiyong Lu, PubMed and beyond: a survey of web tools for searching biomedical literature, Database, Volume 2011, 2011, baq036, https://doi.org/10.1093/database/baq036
Close - Share Icon Share
Abstract
The past decade has witnessed the modern advances of high-throughput technology and rapid growth of research capacity in producing large-scale biological data, both of which were concomitant with an exponential growth of biomedical literature. This wealth of scholarly knowledge is of significant importance for researchers in making scientific discoveries and healthcare professionals in managing health-related matters. However, the acquisition of such information is becoming increasingly difficult due to its large volume and rapid growth. In response, the National Center for Biotechnology Information (NCBI) is continuously making changes to its PubMed Web service for improvement. Meanwhile, different entities have devoted themselves to developing Web tools for helping users quickly and efficiently search and retrieve relevant publications. These practices, together with maturity in the field of text mining, have led to an increase in the number and quality of various Web tools that provide comparable literature search service to PubMed. In this study, we review 28 such tools, highlight their respective innovations, compare them to the PubMed system and one another, and discuss directions for future development. Furthermore, we have built a website dedicated to tracking existing systems and future advances in the field of biomedical literature search. Taken together, our work serves information seekers in choosing tools for their needs and service providers and developers in keeping current in the field.
Database URL:http://www.ncbi.nlm.nih.gov/CBBresearch/Lu/search
Introduction and background
Literature search refers to the process in which people use tools to search for literature relevant to their individual needs. In the context of this review, tools are Web-based online systems; literature is limited to the biomedical domain; and typical user information needs include, but are not limited to, finding the bibliographic information about a specific article, or searching for publications pertinent to a specific topic (e.g. a disease). With the ease of Internet access, the amount of biomedical literature in electronic format is on the rise. As a matter of fact, as pointed out in previous work and shown in Figure 1, the size of the bibliome has grown exponentially over the past few years (1). As of 2010, there are over 20-million citations indexed through PubMed, a free Web literature search service developed and maintained by the National Center for Biotechnology Information (NCBI). PubMed is as part of NCBI’s Entrez retrieval system that provides access to a diverse set of 38 databases (2). PubMed currently includes citations and abstracts from over 5000 life science journals for biomedical articles back to 1948. Since its inception, PubMed has served as the primary tool for electronically searching and retrieving biomedical literature. Millions of queries are issued each day by users around the globe (3), who rely on such access to keep abreast of the state of the art and make discoveries in their own fields.
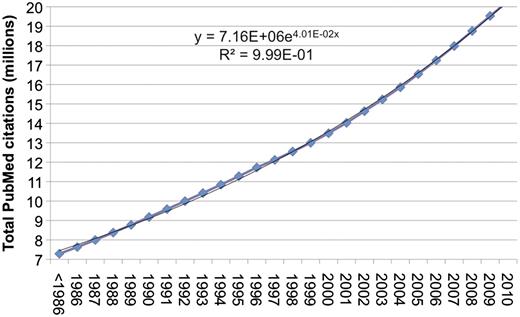
Growth of PubMed citations from 1986 to 2010. Over the past 20 years, the total number of citations in PubMed has increased at a ∼4% growth rate. There are currently over 20-million citations in PubMed. 2010 is partial data (through December 1).
Although PubMed provides a broad, up-to-date and efficient search interface, it has become more and more challenging for its users to quickly identify information relevant to their individual needs, owing mainly to the ever-growing biomedical literature. As a result, users are often overwhelmed by the long list of search results: over one-third of PubMed queries result in 100 or more citations (3). In response to such a problem of information overload, the NCBI has made efforts (see detailed discussion in ‘Changes to PubMed and looking into the future’ section) in enhancing standard PubMed searches by suggesting more specific queries (4). At the same time, the free availability of MEDLNE data and Entrez Programming Utilities (2) make it possible for external entities—from either academia or industry—to create alternative Web tools that are complementary to PubMed.
We present herein a list of 28 such systems, group them by their unique features, compare their differences (with PubMed and one another), and highlight their individual innovations. First and foremost, we aim to provide general readers an overview of PubMed and its recent development, as well as short summaries for other comparable systems that are freely accessible from the Internet. The second objective is to provide researchers, developers and service providers a summary of innovative aspects in recently developed systems, as well as a comparison of different systems. Finally, we have developed a website that is dedicated to online biomedical literature search systems. In addition to the systems discussed in this article, we will keep it updated with new systems so that readers can always be informed of the most current advances in the field.
We believe this work represents the most comprehensive review of systems for seeking information in biomedical literature to date. Unlike many other review articles on text-mining systems (5–11), we limited our focus exclusively to systems that are: (i) for biomedical literature search and (ii) comparable to the PubMed system. The most comparable work is an earlier survey of 18 tools in 2008 (12). However, our review is significantly different in several major aspects. First, the majority of the systems (19/28) in our review were not previously discussed due to different selection criteria or emergence since 2008. Second, we use different classification criteria for categorizing and comparing systems so readers can find discussion from different perspectives. Third, we provide a more detailed overview of each system and its unique features. In particular, we describe PubMed and its recent development in greater detail based on our own experience. Lastly, we have built a website with links to existing systems and mechanisms for registering future systems. All together, our work complements the previous survey, and more importantly it provides one-stop shopping for biomedical literature search systems.
PubMed: the primary tool for searching biomedical literature
Contents and intended audience
PubMed’s intended users include researchers, healthcare professionals and the general public, who either have a need for some specific articles (e.g. search with an article title) or more generally, they search for the most relevant articles pertaining to their individual interests (e.g. information about a disease). A general workflow of how users interact with PubMed is displayed in Figure 2: a user queries PubMed or other similar systems for a particular biomedical information need. Offered a set of retrieved documents, the user can browse the result set and subsequently click to view abstracts or full-text articles, issue a new query, or abandon the current search.
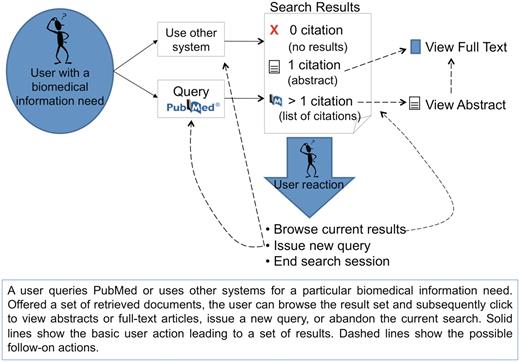
Overview of general user interactions with PubMed (or similar systems) for searching biomedical literature. Adapted from Islamaj Dogan et al., (3).
From a search perspective, PubMed takes as input natural language, free-text keywords and returns a list of citations that match input keywords (PubMed ignores stopwords). Its search strategy has two major characteristics: first, by default it adds Boolean operators into user queries and uses automatic term mapping (ATM). Specifically, the Boolean operator ‘And’ is inserted between multi-term user queries to require retrieved documents to contain all the user keywords. For example, if a user issued the query ‘pubmed search’, the Boolean operator ‘AND’ would be automatically inserted between the two words as ‘pubmed AND search’.
In addition, PubMed automatically compares and maps keywords from a user query to lists of pre-indexed terms (e.g. Medical Subject Headings MeSH®) through its ATM process (http://www.nlm.nih.gov/pubs/techbull/mj08/mj08_pubmed_atm_cite_sensor.html; 13). That is, if a user query can be mapped to one or more MeSH concepts, PubMed will automatically add its MeSH term(s) to the original query. As a result, in addition to retrieving documents containing the query terms, PubMed also retrieves documents indexed with those MeSH terms. Take the earlier example ‘pubmed search’ for illustration, because the word ‘pubmed’ can be mapped to MeSH so the final executed search is [‘pubmed’ (MeSH terms) or ‘pubmed’ (all fields)] and ‘search’ (all fields)’ where the PubMed search tags (all fields) and (MeSH terms) indicate the preceding word will be searched in all indexed fields or only the MeSH indexing field, respectively.
The second major uniqueness of PubMed is its choice for ranking and displaying search results in reverse chronological order. More specifically, PubMed returns matched citations in the time sequence of when they were first entered in PubMed by default. This date is formally termed as the Entrez Date (EDAT) in PubMed.
Other tools comparable to PubMed
Standards for selecting comparable systems
In this work, we selected systems for review based on the following three criteria. First, they should be Web-based and operate on equivalent or similar content as PubMed. Systems that are designed to search beyond abstract, such as full text (e.g. PubMed Central; Google Scholar) or figure/tables [e.g. BioText (14); Yale image finder (15)] are thus not included for consideration in this work. Moreover, we focus on tools developed specifically for the biomedical domain. Hence, some general Web-based services such as Google Scholar are excluded in the discussion. Second, a system should be capable of searching an arbitrary topic in the biomedical literature as opposed to some limited areas. Although most citations in PubMed are of biologically relevant subjects (e.g. gene or disease), the topics in the entire biomedical literature are of a much broader coverage. For example, it includes a number of interdisciplinary subjects such as bioinformatics. In other words, the proposed system needs to be developed generally enough so that different kinds of topics can be searched. Third, the online Web system should require no installation or subscription fee (i.e. freely accessible), which would allow the users to readily experience the service. By these three standards, a total of 28 qualified systems were found and they are listed in Tables 1 and 2 below. Moreover, we classified them into four categories depending on the best match between their most notable features and the category theme. Note that some systems may have features belonging to multiple groups and that within each group, we list systems in reverse chronological order. In Table 1, we show the year when a system was first introduced and highlight major features that distinguish different systems from the technology development perspective. In Table 2, we compare a set of features that affect the value and utility of different tools from a user perspective. For instance, we report the last content update time for each system as most users would like to keep informed with the latest publications. Specifically, we used the PubMed content as the study control and searched for the latest PubMed citation (PMID: 20726112 on 23 August 2010) in all the systems during comparison. When the citation can be found in a system, we consider its content as ‘current’ with PubMed. Otherwise, either an exact date (if such information is provided at the Website) or approximate year is labeled.
| Systems . | Year . | Major features . |
|---|---|---|
| Ranking search results | ||
| RefMed | 2010 | Featuring multi-level relevance feedback for ranking |
| Quertle | 2009 | Allowing searches with concept categories |
| MedlineRanker | 2009 | Finding relevant documents through classification |
| MiSearch | 2009 | Using implicit feedback for improving ranking |
| Hakia | 2008 | Powered by Hakia’s proprietary semantic search technology |
| SemanticMEDLINE | 2008 | Powered by cognition’s proprietary search technology |
| MScanner | 2008 | Finding relevant documents through classification |
| eTBLAST | 2007 | Finding documents similar to input text |
| PubFocus | 2006 | Sorting by impact factor and citation volume |
| Twease | 2005 | Query expansion with relevance ranking technique |
| Clustering results into topics | ||
| Anne O’Tate | 2008 | Clustering by important words, topics, journals, authors, etc. |
| McSyBi | 2007 | Clustering by MeSH or UMLS concepts |
| GoPubMed | 2005 | Clustering by MeSH or GO terms |
| ClusterMed | 2004 | Clustering by MeSH, title/abstract, author, affiliation, or date |
| XplorMed | 2001 | Clustering by extracted keywords from abstracts |
| Extracting and displaying semantics and relations | ||
| MedEvi | 2008 | Providing textual evidence of semantic relations in output |
| EBIMed | 2007 | Displaying proteins, GO annotations, drugs and species |
| CiteXplore | 2006 | EBI’s tool for integrating biomedical literature and data |
| MEDIE | 2006 | Extracting text fragments matching queried semantics |
| PubNet | 2005 | Visualizing literature-derived network of bio-entities |
| Improving search interface and retrieval experience | ||
| iPubMed | 2010 | Allow fuzzy search and approximate match |
| PubGet | 2007 | Retrieving results in PDFs |
| BabelMeSH | 2006 | Multi-language search interface |
| HubMed | 2006 | Export data in multiple format; visualization; etc |
| askMEDLINE | 2005 | Converting questions into formulated search as PICO |
| SLIM | 2005 | Slider interface for PubMed searches |
| PICO | 2004 | Search with patient, intervention, comparison, outcome |
| PubCrawler | 1999 | Alerting users with new articles based on saved searches |
| Systems . | Year . | Major features . |
|---|---|---|
| Ranking search results | ||
| RefMed | 2010 | Featuring multi-level relevance feedback for ranking |
| Quertle | 2009 | Allowing searches with concept categories |
| MedlineRanker | 2009 | Finding relevant documents through classification |
| MiSearch | 2009 | Using implicit feedback for improving ranking |
| Hakia | 2008 | Powered by Hakia’s proprietary semantic search technology |
| SemanticMEDLINE | 2008 | Powered by cognition’s proprietary search technology |
| MScanner | 2008 | Finding relevant documents through classification |
| eTBLAST | 2007 | Finding documents similar to input text |
| PubFocus | 2006 | Sorting by impact factor and citation volume |
| Twease | 2005 | Query expansion with relevance ranking technique |
| Clustering results into topics | ||
| Anne O’Tate | 2008 | Clustering by important words, topics, journals, authors, etc. |
| McSyBi | 2007 | Clustering by MeSH or UMLS concepts |
| GoPubMed | 2005 | Clustering by MeSH or GO terms |
| ClusterMed | 2004 | Clustering by MeSH, title/abstract, author, affiliation, or date |
| XplorMed | 2001 | Clustering by extracted keywords from abstracts |
| Extracting and displaying semantics and relations | ||
| MedEvi | 2008 | Providing textual evidence of semantic relations in output |
| EBIMed | 2007 | Displaying proteins, GO annotations, drugs and species |
| CiteXplore | 2006 | EBI’s tool for integrating biomedical literature and data |
| MEDIE | 2006 | Extracting text fragments matching queried semantics |
| PubNet | 2005 | Visualizing literature-derived network of bio-entities |
| Improving search interface and retrieval experience | ||
| iPubMed | 2010 | Allow fuzzy search and approximate match |
| PubGet | 2007 | Retrieving results in PDFs |
| BabelMeSH | 2006 | Multi-language search interface |
| HubMed | 2006 | Export data in multiple format; visualization; etc |
| askMEDLINE | 2005 | Converting questions into formulated search as PICO |
| SLIM | 2005 | Slider interface for PubMed searches |
| PICO | 2004 | Search with patient, intervention, comparison, outcome |
| PubCrawler | 1999 | Alerting users with new articles based on saved searches |
Within each group, systems are sorted in reverse chronological order.
| Systems . | Year . | Major features . |
|---|---|---|
| Ranking search results | ||
| RefMed | 2010 | Featuring multi-level relevance feedback for ranking |
| Quertle | 2009 | Allowing searches with concept categories |
| MedlineRanker | 2009 | Finding relevant documents through classification |
| MiSearch | 2009 | Using implicit feedback for improving ranking |
| Hakia | 2008 | Powered by Hakia’s proprietary semantic search technology |
| SemanticMEDLINE | 2008 | Powered by cognition’s proprietary search technology |
| MScanner | 2008 | Finding relevant documents through classification |
| eTBLAST | 2007 | Finding documents similar to input text |
| PubFocus | 2006 | Sorting by impact factor and citation volume |
| Twease | 2005 | Query expansion with relevance ranking technique |
| Clustering results into topics | ||
| Anne O’Tate | 2008 | Clustering by important words, topics, journals, authors, etc. |
| McSyBi | 2007 | Clustering by MeSH or UMLS concepts |
| GoPubMed | 2005 | Clustering by MeSH or GO terms |
| ClusterMed | 2004 | Clustering by MeSH, title/abstract, author, affiliation, or date |
| XplorMed | 2001 | Clustering by extracted keywords from abstracts |
| Extracting and displaying semantics and relations | ||
| MedEvi | 2008 | Providing textual evidence of semantic relations in output |
| EBIMed | 2007 | Displaying proteins, GO annotations, drugs and species |
| CiteXplore | 2006 | EBI’s tool for integrating biomedical literature and data |
| MEDIE | 2006 | Extracting text fragments matching queried semantics |
| PubNet | 2005 | Visualizing literature-derived network of bio-entities |
| Improving search interface and retrieval experience | ||
| iPubMed | 2010 | Allow fuzzy search and approximate match |
| PubGet | 2007 | Retrieving results in PDFs |
| BabelMeSH | 2006 | Multi-language search interface |
| HubMed | 2006 | Export data in multiple format; visualization; etc |
| askMEDLINE | 2005 | Converting questions into formulated search as PICO |
| SLIM | 2005 | Slider interface for PubMed searches |
| PICO | 2004 | Search with patient, intervention, comparison, outcome |
| PubCrawler | 1999 | Alerting users with new articles based on saved searches |
| Systems . | Year . | Major features . |
|---|---|---|
| Ranking search results | ||
| RefMed | 2010 | Featuring multi-level relevance feedback for ranking |
| Quertle | 2009 | Allowing searches with concept categories |
| MedlineRanker | 2009 | Finding relevant documents through classification |
| MiSearch | 2009 | Using implicit feedback for improving ranking |
| Hakia | 2008 | Powered by Hakia’s proprietary semantic search technology |
| SemanticMEDLINE | 2008 | Powered by cognition’s proprietary search technology |
| MScanner | 2008 | Finding relevant documents through classification |
| eTBLAST | 2007 | Finding documents similar to input text |
| PubFocus | 2006 | Sorting by impact factor and citation volume |
| Twease | 2005 | Query expansion with relevance ranking technique |
| Clustering results into topics | ||
| Anne O’Tate | 2008 | Clustering by important words, topics, journals, authors, etc. |
| McSyBi | 2007 | Clustering by MeSH or UMLS concepts |
| GoPubMed | 2005 | Clustering by MeSH or GO terms |
| ClusterMed | 2004 | Clustering by MeSH, title/abstract, author, affiliation, or date |
| XplorMed | 2001 | Clustering by extracted keywords from abstracts |
| Extracting and displaying semantics and relations | ||
| MedEvi | 2008 | Providing textual evidence of semantic relations in output |
| EBIMed | 2007 | Displaying proteins, GO annotations, drugs and species |
| CiteXplore | 2006 | EBI’s tool for integrating biomedical literature and data |
| MEDIE | 2006 | Extracting text fragments matching queried semantics |
| PubNet | 2005 | Visualizing literature-derived network of bio-entities |
| Improving search interface and retrieval experience | ||
| iPubMed | 2010 | Allow fuzzy search and approximate match |
| PubGet | 2007 | Retrieving results in PDFs |
| BabelMeSH | 2006 | Multi-language search interface |
| HubMed | 2006 | Export data in multiple format; visualization; etc |
| askMEDLINE | 2005 | Converting questions into formulated search as PICO |
| SLIM | 2005 | Slider interface for PubMed searches |
| PICO | 2004 | Search with patient, intervention, comparison, outcome |
| PubCrawler | 1999 | Alerting users with new articles based on saved searches |
Within each group, systems are sorted in reverse chronological order.
| Systems . | Content last update . | Service provider profile . | Source code available . | System output format . | PubMed ID links . | Full-text links . | Related article links . | Export search results . |
|---|---|---|---|---|---|---|---|---|
| RefMed | 2010 | Academic | × | List | ✓ | × | × | × |
| Quertle | 2010 | Private | × | List | ✓ | ✓ | × | ✓ |
| MedlineRanker | Current | Academic | × | List | ✓ | × | × | × |
| MiSearch | Current | Academic | × | List | ✓ | × | × | × |
| Hakia | 2010 | Private | × | List | ✓ | × | × | × |
| SemanticMEDLINE | 8 June 2010 | Private | × | List | ✓ | × | × | × |
| MScanner | 2007 | Academic | ✓ | List | ✓ | × | × | × |
| eTBLAST | 2010 | Academic | × | List | ✓ | × | × | × |
| PubFocus | Current | Private | × | List | × | × | × | × |
| Twease | Current | Academic | ✓ | List | ✓ | × | ✓ | × |
| Anne O’Tate | Current | Academic | × | List | ✓ | × | ✓ | × |
| McSyBi | Current | Academic | × | List | ✓ | × | × | × |
| GoPubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| ClusterMed | Current | Private | × | List | ✓ | × | × | ✓ |
| XplorMed | Current | Academic | × | List | ✓ | × | × | × |
| MedEvi | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| EBIMed | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| CiteXplore | Current | Govn’t | × | List | ✓ | ✓ | × | ✓ |
| MEDIE | 12 October 2009 | Academic | × | List | ✓ | × | × | × |
| PubNet | Current | Academic | × | Graph | ✓ | × | × | ✓ |
| iPubMed | Current | Academic | × | List | ✓ | × | × | × |
| PubGet | Current | Private | × | List | ✓ | ✓ | × | ✓ |
| BabelMeSH | 2010 | Govn’t | × | List | ✓ | ✓ | × | × |
| HubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| askMEDLINE | 2010 | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| SLIM | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PICO | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PubCrawler | Current | Academic | × | List | ✓ | × | ✓ | ✓ |
| Systems . | Content last update . | Service provider profile . | Source code available . | System output format . | PubMed ID links . | Full-text links . | Related article links . | Export search results . |
|---|---|---|---|---|---|---|---|---|
| RefMed | 2010 | Academic | × | List | ✓ | × | × | × |
| Quertle | 2010 | Private | × | List | ✓ | ✓ | × | ✓ |
| MedlineRanker | Current | Academic | × | List | ✓ | × | × | × |
| MiSearch | Current | Academic | × | List | ✓ | × | × | × |
| Hakia | 2010 | Private | × | List | ✓ | × | × | × |
| SemanticMEDLINE | 8 June 2010 | Private | × | List | ✓ | × | × | × |
| MScanner | 2007 | Academic | ✓ | List | ✓ | × | × | × |
| eTBLAST | 2010 | Academic | × | List | ✓ | × | × | × |
| PubFocus | Current | Private | × | List | × | × | × | × |
| Twease | Current | Academic | ✓ | List | ✓ | × | ✓ | × |
| Anne O’Tate | Current | Academic | × | List | ✓ | × | ✓ | × |
| McSyBi | Current | Academic | × | List | ✓ | × | × | × |
| GoPubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| ClusterMed | Current | Private | × | List | ✓ | × | × | ✓ |
| XplorMed | Current | Academic | × | List | ✓ | × | × | × |
| MedEvi | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| EBIMed | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| CiteXplore | Current | Govn’t | × | List | ✓ | ✓ | × | ✓ |
| MEDIE | 12 October 2009 | Academic | × | List | ✓ | × | × | × |
| PubNet | Current | Academic | × | Graph | ✓ | × | × | ✓ |
| iPubMed | Current | Academic | × | List | ✓ | × | × | × |
| PubGet | Current | Private | × | List | ✓ | ✓ | × | ✓ |
| BabelMeSH | 2010 | Govn’t | × | List | ✓ | ✓ | × | × |
| HubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| askMEDLINE | 2010 | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| SLIM | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PICO | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PubCrawler | Current | Academic | × | List | ✓ | × | ✓ | ✓ |
Tools are listed in the same order as they appear in Table 1. PubMed was used as the study control (assessed on 23 August 2010) for content last update (i.e. current means its content is current with the PubMed content). Latest year information was used when no exact date can be determined. Symbol ✓ stands for yes, and × for no. Govn’t, government.
| Systems . | Content last update . | Service provider profile . | Source code available . | System output format . | PubMed ID links . | Full-text links . | Related article links . | Export search results . |
|---|---|---|---|---|---|---|---|---|
| RefMed | 2010 | Academic | × | List | ✓ | × | × | × |
| Quertle | 2010 | Private | × | List | ✓ | ✓ | × | ✓ |
| MedlineRanker | Current | Academic | × | List | ✓ | × | × | × |
| MiSearch | Current | Academic | × | List | ✓ | × | × | × |
| Hakia | 2010 | Private | × | List | ✓ | × | × | × |
| SemanticMEDLINE | 8 June 2010 | Private | × | List | ✓ | × | × | × |
| MScanner | 2007 | Academic | ✓ | List | ✓ | × | × | × |
| eTBLAST | 2010 | Academic | × | List | ✓ | × | × | × |
| PubFocus | Current | Private | × | List | × | × | × | × |
| Twease | Current | Academic | ✓ | List | ✓ | × | ✓ | × |
| Anne O’Tate | Current | Academic | × | List | ✓ | × | ✓ | × |
| McSyBi | Current | Academic | × | List | ✓ | × | × | × |
| GoPubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| ClusterMed | Current | Private | × | List | ✓ | × | × | ✓ |
| XplorMed | Current | Academic | × | List | ✓ | × | × | × |
| MedEvi | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| EBIMed | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| CiteXplore | Current | Govn’t | × | List | ✓ | ✓ | × | ✓ |
| MEDIE | 12 October 2009 | Academic | × | List | ✓ | × | × | × |
| PubNet | Current | Academic | × | Graph | ✓ | × | × | ✓ |
| iPubMed | Current | Academic | × | List | ✓ | × | × | × |
| PubGet | Current | Private | × | List | ✓ | ✓ | × | ✓ |
| BabelMeSH | 2010 | Govn’t | × | List | ✓ | ✓ | × | × |
| HubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| askMEDLINE | 2010 | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| SLIM | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PICO | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PubCrawler | Current | Academic | × | List | ✓ | × | ✓ | ✓ |
| Systems . | Content last update . | Service provider profile . | Source code available . | System output format . | PubMed ID links . | Full-text links . | Related article links . | Export search results . |
|---|---|---|---|---|---|---|---|---|
| RefMed | 2010 | Academic | × | List | ✓ | × | × | × |
| Quertle | 2010 | Private | × | List | ✓ | ✓ | × | ✓ |
| MedlineRanker | Current | Academic | × | List | ✓ | × | × | × |
| MiSearch | Current | Academic | × | List | ✓ | × | × | × |
| Hakia | 2010 | Private | × | List | ✓ | × | × | × |
| SemanticMEDLINE | 8 June 2010 | Private | × | List | ✓ | × | × | × |
| MScanner | 2007 | Academic | ✓ | List | ✓ | × | × | × |
| eTBLAST | 2010 | Academic | × | List | ✓ | × | × | × |
| PubFocus | Current | Private | × | List | × | × | × | × |
| Twease | Current | Academic | ✓ | List | ✓ | × | ✓ | × |
| Anne O’Tate | Current | Academic | × | List | ✓ | × | ✓ | × |
| McSyBi | Current | Academic | × | List | ✓ | × | × | × |
| GoPubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| ClusterMed | Current | Private | × | List | ✓ | × | × | ✓ |
| XplorMed | Current | Academic | × | List | ✓ | × | × | × |
| MedEvi | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| EBIMed | 2010 | Govn’t | × | Table | ✓ | × | × | × |
| CiteXplore | Current | Govn’t | × | List | ✓ | ✓ | × | ✓ |
| MEDIE | 12 October 2009 | Academic | × | List | ✓ | × | × | × |
| PubNet | Current | Academic | × | Graph | ✓ | × | × | ✓ |
| iPubMed | Current | Academic | × | List | ✓ | × | × | × |
| PubGet | Current | Private | × | List | ✓ | ✓ | × | ✓ |
| BabelMeSH | 2010 | Govn’t | × | List | ✓ | ✓ | × | × |
| HubMed | Current | Private | × | List | ✓ | ✓ | ✓ | ✓ |
| askMEDLINE | 2010 | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| SLIM | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PICO | Current | Govn’t | × | List | ✓ | ✓ | ✓ | × |
| PubCrawler | Current | Academic | × | List | ✓ | × | ✓ | ✓ |
Tools are listed in the same order as they appear in Table 1. PubMed was used as the study control (assessed on 23 August 2010) for content last update (i.e. current means its content is current with the PubMed content). Latest year information was used when no exact date can be determined. Symbol ✓ stands for yes, and × for no. Govn’t, government.
Based on the content of both tables, we have the following observations:
The majority (16/28) of systems contains either ‘Pub’ or ‘Med’ in their name, indicating their strong bond to the PubMed system.
All reviewed systems have been developed continuously during the past 10 or so years, starting from the introduction of PubCrawler in 1999 to iPubMed, the newest member in 2010. It is roughly the same period of time that a significant advance and maturity take place in the fields of text mining and Web technology. Many novel techniques in those two fields (e.g. named entity recognition techniques) were driving forces in the development of various systems reviewed in this work.
Most systems were developed by academics researchers. Yet, several systems also came from the private sector (i.e. Hakia, Cognition, ClusterMed, Quertle) or the public sector (e.g. CiteXplore from the European Bioinformatics Institute). In addition to free access (a requirement for all the systems), the source code of two academic systems (MScanner and Twease) are freely available at their websites under the GNU General Public License.
Similar to the general Web search engines such as Google, the presentation of search results in the reviewed tools is primarily list based. For some systems that perform result clustering, the list can be further grouped into different topics. Other output formats include tabular and graph presentations, which are designed for systems that are able to extract and display semantic relations.
Although only few systems offer links to full-text and related articles, and allow export to bibliographic management software after searches (desirable functions in literature search), one can always (except in one system) follow the PubMed link to use those utilities.
When comparing the four different development themes, improving ranking and the user interface seem to be the more popular directions. In the following sections, we describe each of the 28 systems in greater detail.
Ranking search results
PubMed returns search results in reverse chronological order by default. In other words, most recent publications are always returned first. Although returning results by time order has its own advantages, several systems are devoted to seeking alternative strategies in ranking results.
RefMed (16) is a recent development based on both machine learning and information retrieval (IR) techniques. It first retrieves search results based on user queries. Next, it asks for explicit user feedback on relevant documents and uses such information to learn a ranking function by a so-called learning-to-rank algorithm RankSVM (17,18). Subsequently, the learned function ranks retrieval results by relevance in the next iteration.
Quertle (19) is a recent biomedical literature search engine developed by a for-profit private enterprise. Its core concept recognition features allow the users to incorporate concept categories into their searches. For instance, one of their concept categories represents all protein names, thus users can search all specific proteins as a whole. It is also claimed that they extract relationships based on the context for improving text retrieval. However, its details are not clearly described to the public.
MedlineRanker (20) takes as input a set of documents relating to a certain topic, and automatically learns a list of most discriminative words representing that topic based on a Naïve Bayes classifier. Then it can use the learned words to score and rank newly published articles pertaining to the topic.
MiSearch (21) is an online tool that ranks citations by using implicit relevance feedback (22). Unlike RefMed, it uses user clickthrough history as implicit feedback for identifying terms relevant to user’s information need in the form of log likelihood ratios. MEDLINE citations that contain a larger number of such relevant terms would be ranked higher than those with a lesser number of such terms. In their implicit relevance feedback model, they also take the recency effect into consideration.
Hikia (23) offers access to more than 10-million MEDLINE citations through pubmed.hakia.com. Because it is a product of a private company, it is unclear which ranking algorithm is employed in their system, except that it is said of some kind of semantic search technology.
Semantic MEDLINETM (24) was built based on CognitionSearchTM, a system developed by Cognition’s proprietary Semantic NLPTM technology, which incorporates word and phrase knowledge for understanding the semantic meaning of the English language. The Semantic MEDLINE system adds specific vocabularies from biomedicine in order to better understand the domain specific language. Like Hikia, details are not revealed to the public.
MScanner (25) is mostly comparable to MedlineRanker in terms of its functionality. The major difference is that it uses MEDLINE annotations (MeSH and journal identifiers) instead of words (nouns) in the abstract when doing the classification. As a result, Mscanner is able to process documents faster but it cannot process articles with incomplete or missing annotations.
eTBLAST (26) is capable of identifying relevancy by finding documents similar to the input text. Unlike PubMed’s related articles (27) that uses summed weights of overlapping words between two documents, eTBLAST determines text similarity based on word alignment. Thus, abstract-length textual input is superior to short queries in obtaining good results.
PubFocus (28) sorts articles based on a hybrid of domain specific factors for ranking scientific publications: journal impact factor, volume of forward references, reference dynamics, and authors’ contribution level.
Twease (29) was built on the classic Okapi BM25 ranking algorithm (30) with twists such that retrieval performance can be maintained when query terms are automatically expanded through the biomedical thesauri or post-indexing stemming.
Clustering results into topics
The common theme of the five systems in the second group is about categorization of search results, aiming for quicker navigation and easier management of large numbers of returned results. Such a technique is developed to respond to the problem of information overload: users are often overwhelmed by a long list of returned documents. As pointed out in ref. (31), this technique is generally shown to be effective and useful for seeking relevant information from medical journal articles. As discussed in details below, the five systems mainly differ in the manner by which search results are clustered.
Anne O’Tate (32) post-processes retrieved results from PubMed searches and groups them into one of the pre-defined categories: important words, MeSH topics, affiliations, author names, journals and year of publication. Important words have more frequent occurrences in the result subset than in the MEDLINE as a whole, thus they distinguish the result subset from the rest of MEDLINE. Clicking on a given category name will display all articles in that category. To find a article by multiple categories, one can follow the categories progressively (e.g. first restricting results by year of publication, then by journals).
McSyBi (33) presents clustered results in two distinct fashions: hierarchical or non-hierarchical. While the former provides an overview of the search results, the latter shows relationships among the search results. Furthermore, it allows users to re-cluster results by imposing either a MeSH term or ULMS Semantic Type of her research interest. Updated clusters are automatically labeled by relevant MeSH terms and by signature terms extracted from title and abstracts.
GOPubMed (34) was originally designed to leverage the hierarchy in Gene Ontology (GO) to organize search results, thus allowing users to quickly navigate results by GO categories. Recently, it was made capable of sorting results into four top-level categories: what (biomedical concepts), who (author names), where (affiliations and journals) and when (date of publications). In the what category, articles are further sorted according to relevant GO, MeSH or UniProt concepts.
ClusterMed (35) can cluster results in six different ways: (i) title, abstract and MeSH terms (TiAbMh); (ii) title and abstract (TiAb); (iii) MeSH terms (Mh); (iv) author names (Au); (v) affiliations (Ad) and (vi) date of publication (Dp). For example, when clustering results by TiAbMh, both selected words from title/abstract and MeSH terms are used as filters. Like Hakia, ClusterMed is a proprietary product from a commercial company (Vivisimo) that specializes in enterprise search platforms. Thus, how the filters are selected is not known to the public.
XplorMed (36) not only organizes results by MeSH classes, it also allows users to explore the subject and words of interest. Specifically, it first returns a coarse level clustering of results using MeSH, offering an opportunity for users to restrict their search to certain categories of interest. Next, the tool displays keywords in the selected abstracts. At this step, users can choose to either go directly to the next step or start a deeper analysis of the displayed subjects. The former would present chains of closely related keywords, while the latter allows you to explore the relationships between different keywords and their mentions in MEDLINE articles. Finally by selecting one or more chained keywords, the system returns a list of articles ranked by those selected keywords.
Enriching results with semantics and visualization
The five systems in this group aim to analyze search results and present summarized knowledge of semantics (biomedical concepts and their relationships) based on information extraction techniques. They differ in three aspects: (i) the types of biomedical concepts and relations to be extracted; (ii) the computational techniques used for information extraction; and (iii) how they present extraction results.
MedEvi (37) provides 10 concept variables of major biological entities (e.g. gene) to be used in semantic queries such that the search results are bound to the associated biological entities. Additionally, it also prioritizes search results to return first those citations with matching keywords aligned to the order as they occur in original queries.
EBIMED (38) extracts proteins, GO annotations, drugs and species from retrieved documents. Relationships between extracted concepts are identified based on co-occurrence analysis. The overall results are presented in table format.
CiteXplore (39) is a system that combines literature search with text-mining tools in order to provide integrated access to both literature and biological data. In addition to the content of PubMed, it also contains abstract records from patent applications from the Europe Patent office and from the Shanghai Information Center for Life Sciences, Chinese Academy of Sciences. One other feature of CiteXplore is its inclusion of reference citation information.
MEDIE (40) provides semantic search in addition to standard keyword search in the format of (subject, verb, object) and returns text fragments (abstract sentences) that match the queried semantic relations. Its output is based on both syntactic and semantic parses of the abstract sentences. For example, a semantic search such as ‘what causes colon cancer?’ will require the output sentences to match ‘cause’ and ‘colon cancer’ as the event verb and object, respectively.
PubNet (41) stands for Publication Network Graph Utility. It parses the XML output of standard PubMed queries and creates different kinds of networks depending on the type of nodes and edges a user selects. Nodes can be representatives of article, author or some database IDs (e.g. PDB ids) and edges are constructed based on shared authors, MeSH terms or location (articles have identical affiliation zip codes). The graph networks are drawn with the aid of private visualization software.
Improving search interface and retrieval experience
Systems in this group provide alternative interfaces to the standard PubMed searches. They aim to improve the efficiency of literature search and often take advantage of new Web technologies. They feature novel search/retrieval functions that are currently not available through PubMed, which may be preferred by some users in practice.
iPubMed (42) provides an interactive search interface: search as you type. When a user types several characters into the search box, the system will instantly show any citations containing that text so that users may narrow their searches. In addition, the system allows minor spelling errors.
PubGet (43) displays PDFs directly in search results so that users do not have to follow links in PubMed results to PubMed Central or specific journal websites to get PDFs.
Babelmesh (44) provides an interface so that users can search medical terms and phrases in languages other than English. Currently supported languages include Arabic, Chinese, Dutch, etc. A user’s original query is translated into English and then searched for relevant citations.
HubMed (45) uses Web services to provide various functions ranging from those available in PubMed such as date-sorted search results and automatic term expansion, to new features like relevance-ranked search results; clustering and graphical display of related articles; direct export of citation metadata in many formats; linking of keywords to external sources of information; and manual categorization and storage of interesting articles.
askMEDLINE (46,47) is designed for handling user queries in the form of questions or complex phrases in the medical setting. It was originally developed as a tool for parsing clinical questions to automatically complete the patient, intervention, comparison, outcome (PICO) form, but was later launched as a tool for the non-expert medical information seeker owing to its ability to retrieve relevant citations from parsed medical terms.
SLIM (48) is a slider interface for PubMed searches. It features several slide bars to control search limits in a different fashion.
PICO (49) which stands for patient/problem, intervention, comparison and outcome, is a method used for structuring clinical questions. Its search interface is also available on handhelds.
PubCrawler (50,51) checks and emails daily updates in MEDLINE to the pre-specified searches saved by the users.
Other honorable mentions
Several other systems are noteworthy even though they are not listed in Table 1 due to failing to meet one or more of our predefined requirements:
PubMed Assistant (52), AliBaba (53) and PubMed-EX (54) are three non Web-based systems in the PubMed family (disobey selection criterion #1 which requires systems to be Web-based). PubMed assistant belongs to the group of systems for improving usability: it provides useful functions such as keyword highlighting, easy export to citation managers, etc. Both AliBaba and PubMed-EX are geared towards semantic enrichment by identifying gene/protein, disease and other biomedical entities from the text. In addition, AliBaba also presents co-occurrence results in a graph.
iHop (55), Chilibot (56), PolySearch (57) and Semedico (58) are four representative systems that focus on mining associations between special topics (disobey selection criterion #2 which requires systems to handle general topics). iHop and Chilibot limit their mining to identifying genes and proteins in MEDLNE sentences, while PolySearch supports search over a much broader classes (e.g. diseases). Semidico currently indexes only articles in molecular biology (a sub-area in biomedicine); it mines various biomedical concepts (e.g. gene/protein names) from retrieved documents for enabling faceted navigation. Authority (59) is another example of specialized systems. It uses statistical methods to disambiguate author names, thus making it possible for finding articles written by individual authors.
To improve biomedical literature search, other systems such as PubFinder (60) ReleMed (61), MedMiner (62) and PubClust (63,64) have been proposed. Unfortunately, none of these systems was in service when they were tested on 31 May 2010 (disobey selection criterion #3). PubFinder is like MScanner and MedlineRanker in that it was designed to rank documents by relevancy based on an input set of topic-specific documents. Based on the selected abstracts, a list of words pertinent to the topic is automatically calculated, which is subsequently used in selecting documents belonging to the defined topic. Unlike MScanner or MedlineRanker, it finds informative words based on their occurrences in the input and reference set. ReleMed, recently proposed by Siadaty et al. (61), uses sentence-level co-occurrence as a surrogate for the existence of relationships between query words. MedMiner proposes to filter and organize the large amount of search results returned by PubMed, similar to the idea of categorizing search results. Similarly, PubClust was developed on the basis of self-organizing maps (65) to cluster retrieved abstracts in a hierarchical fashion.
Use cases beyond typical PubMed searches
Based on the novel features in each system described above, we show in Figure 3 a list of specific use scenarios that are beyond typical searches in PubMed. Specifically, we first identified a diverse set of 12 use cases, to each of which we further attached applicable systems accordingly. For instance, one can use tools surveyed in this work to search for experts on a specific topic or to visualize search results in networks. Although traditionally PubMed can not meet many of the listed special user needs, its recent development allowed it to perform certain tasks such as identifying similar publications, alerting users with updates and providing feedback in query refinement. More details are presented in ‘Changes to PubMed and looking into the future’ section.
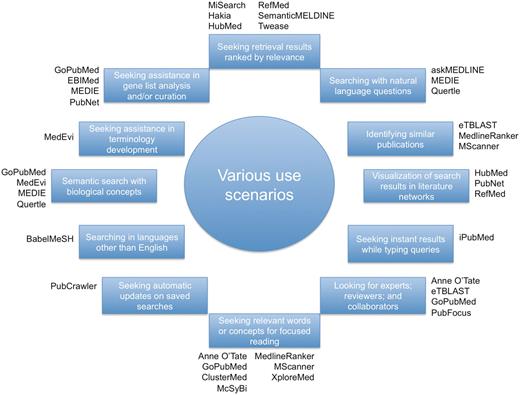
A diverse set of use cases in which different tools may be used.
Discussions on new features
Comparing the 28 systems to PubMed and each other, we see novel proposals for mainly three areas: searching, results analysis and interface/usability.
Searching
Since most users only examine a few returned results on the first result page [Figure 7 in ref. (3)], it is unquestionable that displaying citations by relevance is a desired feature in literature search. The 10 systems listed in ‘Ranking search results’ section differed with PubMed in this regard. Although most of those systems take as input user keywords, they differ from each other on how they process the keywords and subsequently use them to retrieve relevant citations. Like PubMed’s ATM, Twease also has its own query expansion component where additional MeSH terms and others can be added to the original user keywords. This technique can typically boost recall and is especially useful when the original query retrieves few or zero results (13). On the other hand, other systems listed in ‘Ranking search results’ section are mostly aim for improved precision over PubMed’s default reverse time sorting scheme. Their ranking strategies are very different from one another, ranging from traditional IR techniques like explicit/implicit feedback (RefMed/MiSearch) and relevance ranking (Twease), to utilizing domain specific importance factors like journal impact factors and citation numbers (PubFocus), to some unknown proprietary semantic NLP technologies (Hikia and SemanticSearch).
Results analysis
By default, PubMed returns 20 search results in a page and displays the title, abstract and other bibliographic information when a result is clicked. Recent studies focus on two kinds of extensions to the standard PubMed output. First, because a PubMed search typically results in a long list of citations for manual inspection, systems mentioned in ‘Clustering results into topics’ section aim to provide an aid with a short list of major topics summarized from the retrieved articles. Thus, users can navigate and choose to focus on the subjects of interest. This is similar to building filters for the result set (66). In this regard, choosing appropriate topic terms to cluster search results into meaningful groups is the key to the success of such approaches. Currently, most systems rely on selecting either important words from title/abstract or terms from biomedical controlled vocabularies/ontologies (e.g. MeSH) as representative topic terms.
The second extension to the standard PubMed output is due to the advances in text-mining techniques. In particular, semantic annotation is believed to be one of the probable cornerstones in future scientific publishing (67) despite the fact that its full benefits are yet to be determined. Thus with the development and maturity of techniques in named entity recognition and biomedical information extraction, some systems present summarized results of deep semantic enrichment. Existing systems (‘Enriching results with semantics and visualization’ section) have mostly focused on finding genes, proteins, drugs, diseases and species in free text and their biological relationships such as protein–protein interactions. Problems in these areas have received the most attention in the text mining community (68,69).
Interface and usability
In addition to providing improved search quality, a number of systems strive to provide a better search interface, including various changes to input and output. An innovative feature in iPubMed is ‘search-as-you-type’, thus enabling users to dynamically choose queries while inspecting retrieved results. Other proposals for an alternative input interfaces facilitate user-specific questions (PICO, askMedline), allow non-English queries (BabelMeSH), and promote use of sliders to set limits (SLIM). With respect to changes to output, there are two major directions. First, two systems employ additional components to make summarized results visible in graphs (ALiBaba and PubNet). Second, several systems provide easier access to PDFs (PubGet) and external citation mangers (PubMed assistant; HubMed).
Changes to PubMed and looking into the future
In response to the great need and challenge in literature search, PubMed has also gone through a series of significant changes to better serve its users. As shown in Figure 4, many of the recent changes happened during the same time period the 28 reviewed systems were developed. So they may have learned from each other. Indeed, some features were first developed in PubMed (e.g. related articles) while others in third party applications (e.g. email alerts).
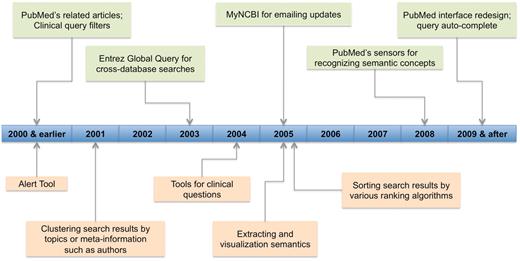
Technology development timeline for PubMed (in light green color) and other biomedical literature search tools (in light orange color). For PubMed, it shows the staring year when various recent changes (limited to those mentioned in ‘Changes to PubMed and looking into the future’ section) were introduced. For other tools, we show the time period in which tools of various features were first appeared.
A new initiative geared towards promoting scientific discoveries was introduced to PubMed a few years ago. Specifically, by providing global search across NCBI’s different databases through the Entrez System (http://www.ncbi.nlm.nih.gov/gquery/), users now have integrated access to all the stored information in different databases to know about a biological entity—be it related publications, DNA sequences or protein structures. Furthermore, inter-database links have been established and made obvious in search result pages, making the related data readily accessible between literature and other NCBI’s biological databases. For instance, through integrated links originating in PubMed results, users can access information about chemicals in PubChem or protein structures in the Structure database. Another category of discovery components is known as sensors (http://www.nlm.nih.gov/pubs/techbull/nd08/nd08_pm_gene_sensor.html; http://www.nlm.nih.gov/pubs/techbull/mj08/mj08_pubmed_atm_cite_sensor.html). A sensor detects certain types of search terms and provides access to relevant information other than literature. For instance, PubMed’s gene sensor detects gene mentions in user queries and shows links directing users to the associated gene records in Entrez Gene. Although these new additions are specific to PubMed and developed independently, they nevertheless all reflect the idea of semantically enriching the literature with biological data of various kinds, to achieve the goal of more efficient acquisition of knowledge.
With respect to research and retrieval, there are also several noteworthy endeavors in PubMed development although its default sorting schema has been kept intact. First, the related article feature was integrated into PubMed so that users can readily examine similar articles in content. eTBLAST has a similar feature, but as explained earlier, the two systems rely on different techniques for obtaining similar documents. Second, specific tools were added into PubMed for different information needs. For instance, the citation matcher is designed for those who search for specific articles. Another example is clinical queries, an interface designed to serve the specific needs of clinicians. It is fundamentally akin to the idea of categorizing search results (‘Ranking search results’ section) because the tool essentially discards any non-clinical results using a set of predefined filters. Finally, in order to help users avert a long list of return results and narrow their searches, a new feature named ‘also try’ was recently introduced, which offers query suggestions from the most popular PubMed queries that contain the user search term (4).
Regarding the user interface and usability, the My NCBI tool was introduced to PubMed, which let users select and create filter options, save search results, apply personal preferences like highlighting search terms in results, and share collections of citations. Similar to PubCrawler, it also allows users to set automatic emails for receiving updates of saved searches. Additional search help such as a spell checker and query auto-complete have also been deployed in PubMed. Finally in 2009, the PubMed interface including its homepage was substantially redesigned such that it is now simplified and easier to navigate and use.
Literature search is a fundamentally important problem in research and it will only become harder as the literature grows at a faster speed and broader scope (across the traditional disciplinary boundaries). Therefore we expect continuous developments and new emerging systems in this field. In particular, with the advances in search and Web technologies in general, we are likely to see progress in literature search as well. With the maturity of biomedical text-mining techniques in recognizing biological entities and their relations, better semantic identification and summarization of search results may be achieved, especially for such entities as author names, disorders, genes/proteins and chemicals/drugs as they are repeatedly and heavily sought topics (3,70) in biomedicine. In addition, one key factor for future system developers is the need to keep their content current with the growth of the literature, as literature search has a recency effect—most users still prefer to be informed of the most current findings in the literature. Finally, to be able to provide one-stop shopping for all 28 reviewed systems plus the ones in the ‘Other honorable mentions’ section and keep track of future developments in this area, we have built a website at http://www.ncbi.nlm.nih.gov/CBBresearch/Lu/search. It contains for every system, a highlight and short description of its unique features, one or more related publications, and a link to the actual system on the Internet. To facilitate busy scientists to quickly find appropriate tools for their specific search needs, we have built a set of search filters. For instance, one can narrow down the entire list of systems to the only ones that keep its content current with PubMed. Future systems will be added to the website either through our quarterly update or by individual request. On the website, we have set up a mechanism for registering future systems. Once we receive such a request, we will curate the necessary information (e.g. system highlights) about the submitted system and make it immediately available at the website.
Conclusions
By our three selection standards, a total of 28 Web systems were included in this review. They are comparable to PubMed given that they are designed for the same purpose and make use of full or partial PubMed data. We first provided a general description of PubMed including its content and unique characteristics. Next, according to their different features, we classified the 28 systems into four major groups in which we further described each of them in greater detail and showed their differences. Finally we reviewed the 28 systems as a whole and discussed their innovative aspects with respect to searching, result analysis and enrichment, and user interface/usability. This review can directly serve both non-experts and expert users when they wish to find systems other than PubMed. Moreover, the review provides a detailed summary for the recent advances in the field of biomedical literature search. This is particularly useful for existing service providers and anyone interested in future development in the field. Finally the constructed website make an integrated and readily access to all reviewed systems and provides a venue for registering future systems.
Acknowledgements
The author is grateful to the helpful discussion with John Wilbur, Minlie Huang and Natalie Xie.
Funding
Funding for this work and open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest: None declared.



