-
PDF
- Split View
-
Views
-
Cite
Cite
Michael Zouberakis, Christina Chandras, Morris Swertz, Damian Smedley, Michael Gruenberger, Jonathan Bard, Klaus Schughart, Nadia Rosenthal, John M. Hancock, Paul N. Schofield, George Kollias, Vassilis Aidinis, Mouse Resource Browser—a database of mouse databases, Database, Volume 2010, 2010, baq010, https://doi.org/10.1093/database/baq010
Close - Share Icon Share
Abstract
The laboratory mouse has become the organism of choice for discovering gene function and unravelling pathogenetic mechanisms of human diseases through the application of various functional genomic approaches. The resulting deluge of data has led to the deployment of numerous online resources and the concomitant need for formalized experimental descriptions, data standardization, database interoperability and integration, a need that has yet to be met. We present here the Mouse Resource Browser (MRB), a database of mouse databases that indexes 217 publicly available mouse resources under 22 categories and uses a standardised database description framework (the CASIMIR DDF) to provide information on their controlled vocabularies (ontologies and minimum information standards), and technical information on programmatic access and data availability. Focusing on interoperability and integration, MRB offers automatic generation of downloadable and re-distributable SOAP application-programming interfaces for resources that provide direct database access. MRB aims to provide useful information to both bench scientists, who can easily navigate and find all mouse related resources in one place, and bioinformaticians, who will be provided with interoperable resources containing data which can be mined and integrated.
Database URL: http://bioit.fleming.gr/mrb
Introduction
The recent successes in decoding the genome of humans and mice reveal that they both code for ∼20 000 genes. Because of the recent divergence of the mouse and human genomes, >99% of human genes have analogues in the mouse. The close homology in sequence extends to function, and many mouse and human homologues have very similar functions. Nevertheless, the role of most of these genes in normal development and physiological processes, as well as their involvement in disease, is poorly understood. The major challenge of the post-genomic era is the attribution of function to genes and pathways, and the use of model organisms such as the mouse to provide phenotype/genotype relations is now established as a key approach to discovering normal gene function.
The numbers of sporadic or targeted mutations in mouse genes have recently been augmented by the activities of the International Mouse Knockout Consortium (IKMC) (1), which, within 2 years, will have knockouts available for all of the genes in the mouse genome. Researchers are increasingly exploiting mouse models to examine the complex mechanisms regulating human disease pathophysiology through the application of functional genomic technologies (2) and mobilisation of this huge resource is now yielding a large volume of rich and novel data (1). Much of this data, together with information on bioresources (Mice and ES cells), is now being shared through online resources that have become indispensable tools for scientists working on gene function and human disease. The propagation of these mouse databases creates a number of new coordination challenges, both technical and conceptual, which need to be met in order to realise the full potential of these global activities. These include uptake of formalized experimental descriptions and data standardization, database interoperability, database visibility outside the context of the local initiative and database financial sustainability. In this context, we describe the Mouse Resource Browser (MRB; http://bioit.fleming.gr/mrb), a database of mouse databases created with the help of the European Commission framework programs MUGEN (Animal models of human immunological diseases; www.mugen-noe.org) and CASIMIR (Coordination and Sustainability of International Mouse Informatics Resources; http://www.casimir.org.uk).
MRB is a resource management project that provides an index of 217 publicly available mouse resources, classified in 22 categories intended for both bench scientists and bioinformaticians. Apart from basic information on database availability and content, MRB provides information on the controlled vocabularies and data standards (ontologies and minimum information standards) used by these resources together with technical information on direct and programmatic access (e.g. web services, BioMart installations), so identifying facilities for data integration and database interoperability. To promote interoperability, MRB has incorporated a modified version of the MOLGENIS bio-software system; this allows automatic generation of re-distributable Java SOAP application-programming interfaces (APIs) for resources that enable direct database access. In order to provide a formal, standardized quality assessment scheme for all recorded resources, MRB uses CASIMIR's Database Description Framework (DDF), a key-point based summary.
Database design, implementation and accessibility
MRB is the front-end of a relational, fully normalized PostgreSQL database, and is a typical Java EE application that follows the MVC architectural pattern, generating three transparent layers: the Enterprise JavaBeans (EJB) layer, the intermediate Session layer and the interface/web layer. The database's schema has been kept as simple as possible and has avoided the extended use of stored procedures and database-management-system (DBMS) specific functions and types in an attempt to keep the application DBMS agnostic. The EJB layer, an object-oriented (OO) API mapped to and in harmony with the design philosophy of the database, has been kept simple. The complexity of most of the relational and combinatorial functionality is handled by the intermediate layer, while the interface layer handles data representation. MRB is currently deployed on Sun’s open-source Glassfish application server. Basic information on database development and implementation, including a schematic diagram illustrating its architecture (Figure 1), can be found on MRB’s ‘About’ page. The source code of MRB is available under the GNU general public licence (GPL) as a binary download and via cvs from the CASIMIR sourceforge project page (http://sourceforge.net/projects/casimir-org-uk/). All data in MRB are freely available to interested users through downloadable weekly database dumps. Programmatic data access is enabled via SOAP web services. Database dumps and web service access details can be found on the ‘Data Access’ page of MRB.
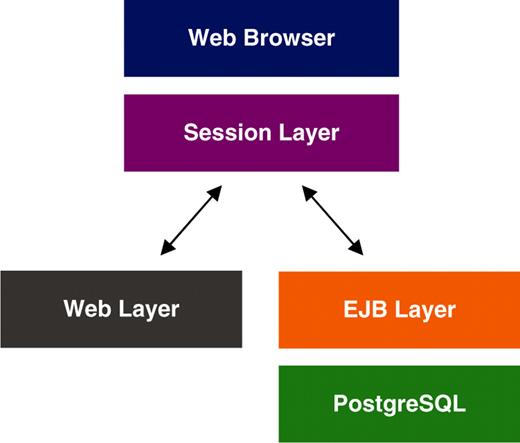
Content management
MRB’s data collection was compiled and is being updated through extensive literature review, web browsing, direct contact with resource personnel, via MRB’s online questionnaire (http://bioit.fleming.gr/mrb/Controller?workflow=imouse), as well as by user recommendations. This questionnaire (for responses, see below) addresses both technical issues and DDF-derived criteria and has highlighted important usage statistics on ontologies and minimum information standards by the international community, presented elsewhere (3). Similarly, MRB’s questionnaire has also allowed valuable conclusions on the financial sustainability models of databases, presented in a separate report (4).
In order to keep MRB up-to-date, its content is regularly updated by curators that annually contact each resource requesting additional or altered information on the existing entry in MRB (resource pages carry update information). These updates are made by MRB’s curation team, who carefully check collected or submitted data for accuracy and completeness. MRB also informs the user of whether the data presented have been provided by the resource itself or by the MRB curation team through literature and web searches. Although MRB can support multiple user groups with different levels of access rights in an ordinary content management system (CMS) fashion, it currently allows restriction-free read access to all visitors.
Content delivery
The easiest way to query the database is by formulating case insensitive free text queries. Users can type the desired query words (or phrases within double quotes) into the text box provided at the top of each MRB page. Returned results by default contain all key words and may include mouse resources, resource categories, ontologies and minimum information standards. Complex queries can be performed through the advanced search page, which incorporates a free text box and refining options for database types, resource categories, programmatic access methods, ontologies and minimum information checklists. Similarly, MRB provides a browsing/filtering interface to the underlying data, allowing formulation of queries that coordinate screening of certain biological databases and resources. In addition, the mouse resource can be browsed using two dropdown menus on the index page; one to sort the collection alphabetically or chronologically and a second one to filter data according to their resource category.
MRB aims to serve both bench scientists and bioinformaticians. The former can retrieve a comprehensive list of online resources and databases pertaining to the laboratory mouse into one resource. This can be achieved via (i) the alphabetical or accession date listing of all 217 resources, (ii) a list of categories which directs the user to the list of resources under the particular category or (iii) more specialised searches through the ‘Search’ box or ‘Advanced Search’ button. Bioinformaticians can in addition use the ‘Technical’ tab in each resource to obtain all the information needed to permit data extraction from that resource and to have programmatic data access where available. Finally MRB’s Data Access capabilities and additional technical information can be found under the ‘Data Access’ tab.
Content structure
MRB currently lists 217 online mouse resources, classified according to their content in 22 categories and accessible through the ‘Resources/Categories’ button via MRB’s top menu. The list is interactive and additional information such as the number of resources classified under the particular category as well as a short text describing the type of resources recorded is also available.
The mouse resource is the application's fundamental entity and its data set is covered by four sections which are accessed by tabs (Figure 2).
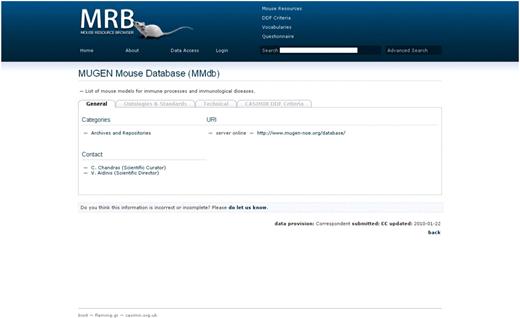
Screen shot of MRB; the view of a mouse resource demonstrating the use of tabs per data set section. Here the ‘General’ tab is on display, which includes a short description of the resource, the categories under which the particular resource is assigned, the URL(s) of the particular database with an interactive link provided and an interactive contact for users to get in touch with the personnel of the particular resource.
The General section/tab
This presents the general information on a biological resource. This includes a short text description of the resource, a list of the content-based categories the resource falls into, the URL(s) of the particular database and contacts for the user to get in touch with the personnel of the particular resource.
The Ontologies & Standards section/tab
This lists those ontologies and minimum information for biological and biomedical investigations (MIBBI) standards adopted by each resource. The scientific value of any dataset is greatly enhanced if it is annotated with a widely used controlled vocabulary or ontology as these facilitate interoperability. The popularity of the approach has led to a proliferation of ontologies, and in turn to the creation of the Open Biomedical Ontologies (OBO) consortium which has created an evolving set of shared principles that coordinate the adaptation and development of existing ontologies (5). The ontology in use by a resource (if any) is hyperlinked to a page/dataset consisting of a short description, a record of whether the ontology is listed in the OBO Foundry and an indication of whether it is non-OBO compliant and only implemented locally. A list of external links to appropriate resources is also provided (e.g. the ontology's homepage and its latest downloadable version in OBO and/or OWL format). All ontologies are fully searchable and indexed in an Ontologies list, accessible through the ‘Vocabularies/Ontologies’ button via MRB's top menu.
In parallel with ontologies and OBO, the MIBBI project fosters coordinated development of extant minimum information checklists needed to fully understand the context, methods, data and conclusions that pertain to an experiment (6). The most well known MIBBI project is still the original MIAME (minimum information about a microarray experiment) checklist (7), a current requirement for the publication of microarray data in many journals. As with ontologies, the Ontologies & Standards section/tab indicates any MIBBI protocol(s) used by the resource, followed by a short description and links. An index of fully searchable and manageable MIBBI projects is accessible through the ‘Vocabularies/MIBBI’ button on MRB's top menu.
The Technical section/tab
This holds technical information focusing on implementation details and instructions for programmatic-access. The mouse resources are split into three categories that include relational databases, object-oriented databases and flat files. Under the ‘Implementation’ heading, the technical tab indicates the category that each resource belongs to, lists the programming languages and the database management system(s) used to develop the resource and the server technology on which it is deployed. Additionally, any available file downloads related to the resource’s schema, such as complete or partial database dumps, images with diagrams modelling the schema or hyperlinks to database dump repositories, can be found on this tab under the ‘Dumps & Files’ heading.
More importantly, MRB provides information on the various programmatic access methods for each resource. This set of information includes any links to web pages describing how to access a resource programmatically, links to BioMart query interfaces and direct links to Web Service Description Language (WSDL) files.
All resources under the Web Service Access subset of the technical tab are characterized with an additional indicator reflecting the status of the server to which each link points and a web service analysis servlet, entitled ‘wsAnalyzer’; its role is to break down WSDL documents on the fly, detect the methods that enable remote access and analyze their input and output parameters. Findings are stored in MRB's database and subsequently presented in a human-readable format. MRB’s aim here is to assist users in deciphering SOAP web service descriptors, but the premise of collecting analyzed WSDL information is to detect commonalities on which web service standardization can be built. The current version of MRB's wsAnalyzer can only process WSDL 1.1 and 2.0 compliant SOAP web service endpoints; possible support for Web Application Description Language (WADL) (8) is under discussion.
In an attempt to provide a convenient SOAP web service generation tool for the mouse community, the open source MOLGENIS biosoftware project (9) has been slightly modified and incorporated into MRB, so enabling automatic generation of standard Java SOAP APIs to databases that allow direct database access. These MOLGENIS-derived functionalities comprise MRB's ‘wsGenerator’ that can be found under the ‘Direct Database Access’ subset of the technical tab. wsGenerator is not restricted to MRB's and is available through http://bioit.fleming.gr/molgenis-ng. Resources interested in participating in this scheme only need to enable remote access to their database: this opens a port for a dedicated database user with minimum access rights (read permissions). Once the server's host-name, the port, user-name and password are provided, the wsGenerator can, on request, parse the database schema and produce: (i) a MOLGENIS XML file modelling all database entities including all their fields and constraints, (ii) an Entity-Relationship (ER) diagram of the database in png image format, (iii) a set of Java wrapper classes for each table in the database and (iv) a Java SOAP web service class to programmatically access the data.
It is important to note that the information presented on the technical tab of MRB appears under-curated for most indexed resources. Based on MRB's collection of resources, it is evident that most resources within the mouse community do not offer direct database access, programmatic access to their data or alternative methods of obtaining them (e.g. providing regular downloadable database dumps). It should be noted that, despite efforts by, and on behalf of, MRB's curation team, attempts to collect information on implementation details were not always successful as some resource personnel were reluctant or, occasionally, lacked the expertise to provide the requested information.
The CASIMIR DDF Criteria section/tab
This provides users with a ready summary of the resource. The DDF criteria focus on specific topics or areas of importance (quality and consistency, currency, accessibility, output, technical documentation, data representation standards, data structure standards, user support and versioning) and have three different levels of maturity. DDF criteria (accessible through the relevant button at the top menu) were established by the CASIMIR consortium with the aim of standardizing database descriptions (further allowing quick benchmarking and loose evaluation) thus facilitating the choice of a resource for a given task, as well as its integration.
Questionnaire responses
MRB includes an online questionnaire that allows resource personnel to individually fill in and indirectly ‘self-curate’ relevant information about their resource. The questionnaire includes six tabbed sets of questions on basic information, resource description, curation & updates, data structure & vocabularies, database sustainability, and computational information. Most questions have checklist answers so that the user is only asked to enter new text where it really is required. The aim is to make the questionnaire as user-friendly as possible. Users may click ‘submit’ at any time and send the relevant information for the MRB staff to use the responses to update the resource’s data. Should there be any discrepancies or missing information MRB staff immediately contact the respective resource and requests the required clarifications. MRB has contacted each resource individually via email asking them to fill in this online questionnaire and 79 out of 212 (37%) resources responded to the questionnaire, 51% being of European origin, 38% from the United States of America and the remaining 11% from Canada, Japan and Australia (Figure 3). MRB staff has used this information both to curate the resources in MRB accurately, and to examine the evolving database and resource landscape.
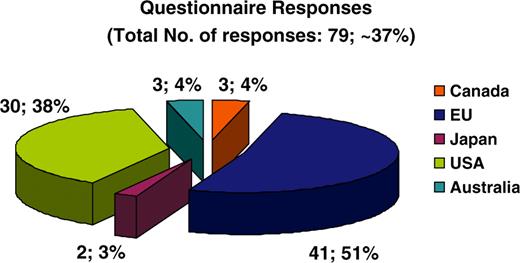
Pie chart depicting the overall online questionnaire responses obtained. Fifty-one percent of responses originated from European countries, 38% from the USA, 3% from Japan and finally 4% each from Canada and Australia.
With regard to curatorial information, of the 28 resources that answered the respective questions, 43% are updated on a monthly basis, 46% annually, while the remaining 11% corresponds to decommissioned resources, meaning that the information is still available online but is no longer updated (Figure 4A). The majority of resources (73%) are manually curated, 8% are automatically curated while 19% have no explicit consistency assurance with regard to the information displayed (Figure 4B). Following the clear need for use of ontologies in the biomedical domain and the notable work performed by the OBO foundry, it was expected that the majority of databases would be using OBO ontologies. It turns out that of the 36 resources that answered the particular question, 72% do use OBO ontologies of which 19% use PATO and the remaining 81% use other OBO ontologies (Figure 5). With regard to database accessibility as identified by the 25 resources that provided related information, 52% of resource data may be obtained via both a web browser interface and another programmatic access (i.e. WebServices, Biomart, etc.), 16% of resources have their data accessed via a web browser interface in addition to public data dumps, while 32% of databases just allow browser access (Figure 6A). Of the databases that provide additional programmatic access (10 respondees), 42% have developed WebServices and 25% show preference for using BioMart (Figure 6B). MRB has also collected some interesting information with regard to the initial funding acquired for the creation of biological databases and resources in addition to funds for their long-term financial maintenance. These responses have been extensively discussed by Chandras et al. (4).
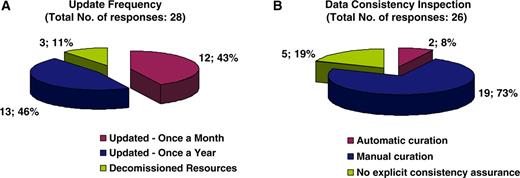
Pie charts representing the curatorial information for each biological database and resource. Forty-three percent of resources are updated on a monthly basis, 46% annually and the remaining 11% corresponds to resources that have become decommissioned (A). Seventy-three percent of resources are curated manually, 8% use an automatic curation system, and 19% of resources have no explicit way of assuring data consistency with regard to the displayed information (B).
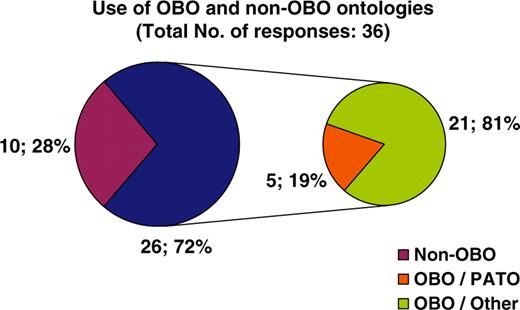
Pie chart illustrating the use of ontologies by biological databases. Twenty-eight percent of resources do not use any ontology for their data description. Seventy-two percent of resources use an ontology developed by the OBO foundry, of which 19% use PATO and 81% another OBO ontology.
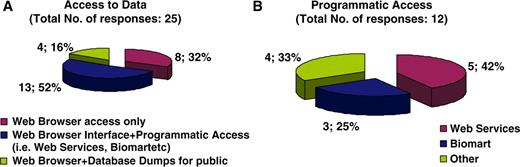
Pie chart representing database accessibility. Fifty-two percent of resource data may be obtained via both a web browser interface in addition to another programmatic access (i.e. WebServices, Biomart, etc.), 16% of resources have their data accessed via a web browser interface and public data dumps, while the remaining 32% of databases allow data access simply on web Browser access (A). Of the databases that provide additional programmatic access, other than a web browser interface, 42% of resources have developed WebServices and 25% show preference on using BioMart (B).
Discussion
The MRB, in addition to being a content-management system for mouse online resources and databases, aims to become an intermediary link between resources for the mouse community. Focusing on integration and interoperability, the collected information includes technical and programmatic accessibility details together with ontologies and minimum-information checklists. The use of standards for both ontologies and metadata figuration is considered essential for the integrational potential of resources to be realized, and is slowly becoming common practice within the mouse community, but more needs to be done if mouse resources are to be made interoperable. While most of the indexed resources provide minimal programmatic access to their data, some curators were reluctant to provide technical information. It is of course a concern for the community that resources failing to keep pace with current and future technological developments may degrade the value of their data. Review of interoperability technologies and the use-case example developed by CASIMIR members (10) pinpoints future directions and demonstrates how technologies like web services (11) and software like Taverna (12) and MOLGENIS (9) can be utilized to accomplish interoperation. In this context, MRB is collecting and analyzing information on implemented programmatic access methods via the wsAnalyzer, and it can also act as a web-service-application server for resources via the wsGenerator.
By providing both web-service compositional and de-compositional tools and including an index of most online mouse resources with advanced search facilities, MRB hopes to introduce bench scientists to new online resources and to help scientific programmers develop bioinformatics applications that combine data from multiple resources. In the latter context, MRB could be used to detect resources that use SOAP web services to enable programmatic access to their data and specific OBO ontologies to structure them. Once MRB yields results, users can employ wsAnalyzer to inspect the web service provided by each resource and build their applications around the returned results. In addition, resources that may utilize specific ontologies of interest to the user but not provide web services, could be accessed via the SOAP API generated by wsGenerator. A bioinformatician could thus download the generated Java classes and incorporate them into an application. Finally, we point out that MRB can easily be customized by replacing the mouse-specific categories, so allowing it to be used by any bio-community as an advanced content management tool for resources.
Funding
Sixth Research Framework Programme of the European Union (CASIMIR LSH-2005-1.1.0-1, MUGEN LSHGCT-2005-005203). Funding for open access charges: CASIMIR LSH-2005-1.1.0-1.
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank all CASIMIR members as well as participants at the various CASIMIR meetings for fruitful discussions.
References
Author notes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.



