-
PDF
- Split View
-
Views
-
Cite
Cite
Christina Chandras, Thomas Weaver, Michael Zouberakis, Damian Smedley, Klaus Schughart, Nadia Rosenthal, John M. Hancock, George Kollias, Paul N. Schofield, Vassilis Aidinis, Models for financial sustainability of biological databases and resources, Database, Volume 2009, 2009, bap017, https://doi.org/10.1093/database/bap017
Close - Share Icon Share
Abstract
Following the technological advances that have enabled genome-wide analysis in most model organisms over the last decade, there has been unprecedented growth in genomic and post-genomic science with concomitant generation of an exponentially increasing volume of data and material resources. As a result, numerous repositories have been created to store and archive data, organisms and material, which are of substantial value to the whole community. Sustained access, facilitating re-use of these resources, is essential, not only for validation, but for re-analysis, testing of new hypotheses and developing new technologies/platforms. A common challenge for most data resources and biological repositories today is finding financial support for maintenance and development to best serve the scientific community. In this study we examine the problems that currently confront the data and resource infrastructure underlying the biomedical sciences. We discuss the financial sustainability issues and potential business models that could be adopted by biological resources and consider long term preservation issues within the context of mouse functional genomics efforts in Europe.
Introduction
In our attempt to better understand the biology of human disease we are generating increasingly diverse and specialized data sets, many of which are extremely large and complex, with the result that when primary data is put in the public domain it is scattered through an increasing number of knowledge domain specific databases and bioresources. These databases contain genomic (including sequencing, expression and microarray), proteomic (structure and function) and metabolomic data as well as information about function, structure, localization and clinical effects of mutations. Furthermore, with the increased attention recently given to mouse mutants that serve as models for human disease and the development of novel therapeutic strategies there has been a proliferation of material resources serving to support mouse research. Information on these material resources is commonly presented through databases such as the International Mouse Strain Resource (IMSR) [1,2]. Biological databases have consequently become an important tool in assisting scientists to understand and explain biological molecules and processes, in addition to their interactions.
Since biological knowledge is distributed worldwide and therefore among many differently specialized databases, it is difficult and frequently impossible to ensure preservation and consistency of information as well as data quality. Currently, much of the collected data are stored in a way that does not always guarantee future retrieval by other researchers [3]. Assured growth, persistence and accessibility of databases are therefore imperative to encourage and support data deposition. Additionally, standardization of data representation and transfer is required for enabling the integration of existing and new databases. At present, biological databases cross-reference other databases with accession numbers or IDs as one way of linking their related knowledge together. Much current European effort is being expended in developing modes of data integration and database interoperability, either as a ‘one-stop-shop’ federation or more recently in the development of ‘smart’ clients which integrate data from multiple sources or run tailor-made workflows [4].
A major problem for most databases is securing financial support for the bioinformaticians and curators who create and maintain them [5,6]. Development and maintenance of databases is a costly activity and while it is hard to generalize about average costs as these will vary depending on the resource's size and complexity, personnel costs in different countries, etc., we can give an example for the European Mouse Mutant Archive (EMMA) database (http://www.emmanet.org/). The informatics for EMMA require three full time equivalent posts and 13% of the overall project funding. Even popular databases commonly lack secure funding and frequently face loss of their original support after a few years in development. Hence, long-term sustainability of databases requires adequate and reliable sources of funding. In this article, we will give an overview of the current financial support situation, potential business models that could be adopted by databases for their long-term financial support, and the attempts that have been made so far.
CASIMIR (http://www.casimir.org.uk), a Coordination Action funded by the European Commission, focuses on the dissemination and integration of databases relevant to the mouse as a model organism for human disease. The overall aim of the project is to identify the factors which inhibit the free flow of data and resources for mouse functional genomics and to determine what is needed to overcome these in order to establish a framework of interoperable databases with concomitant added value to the scientific community. Sustainability is a major challenge and CASIMIR aims to make recommendations to the European Commission and the community on the extent to which databases might become self-sustained in terms of data deposition, usage, development and financial support. CASIMIR will also examine what potential business models could be adopted by biological resources for their financial sustainability and long-term preservation.
Data and biological resources
Publication of experimental results and sharing of the related research materials have long been key elements of the life sciences. Indeed scientific progress depends on the ability of researchers to access and exploit data and materials reported in publications so that they can subsequently build on these findings. Publications also serve as a means of receiving intellectual credit and recognition which subsequently enhance a researcher's career prospects and potential for research support. It is however no longer adequate to share data through traditional modes of publication, and, particularly with high throughput (‘-omics’) technologies, sharing of datasets requires submission to public databases as has long been the case with nucleic acid and protein sequence data. This presents new challenges in extending the traditional publication model to the New Biology.
The traditional quid pro quo arrangement, where authors receive credit and acknowledgements in exchange for disclosure of their scientific findings, has been re-evaluated by a US National Academy of Sciences committee. The responsibility of authors to share data and materials referenced in their publications, the role of journals to impose requirements for data and material sharing, and whether a common set of requirements for sharing should exist has been closely examined and the concept of the ‘uniform principle for sharing integral data and materials expeditiously’ (UPSIDE) [7,8] has been established.
Biological resource centers (BRCs) are centralized repositories that specialize in storing and distributing materials, such as mice or ES cells and their associated data. Both repository and service functions contribute to the needs of national and international consortia, as well as individual laboratories and research institutes in support of academic research programs. A central role for the BRCs is to facilitate the principles set out by UPSIDE and embrace the open-access policy, quality of material, data integration and sustainability. It is crucial that the scientific community, public funding bodies and governments acknowledge these issues as being of primary importance.
In accordance with the aforementioned responsibilities of authors, journals and BRCs came the recently published guidelines by the Organization for Economic Co-operation and Development (OECD) [9] asserting that in order to comply with the data-sharing imperative, adequate and reliable sources of funding are required to facilitate the sharing infrastructure and, as part of that, the long-term stability of BRCs [10]. If, for financial reasons, BRCs are unable to perform their tasks under conditions that meet the requirements of scientific research and the demands of industry, scientists will either see valuable information lost or being transferred into a strictly commercial environment with at least two consequences: (i) blockade of access to this information and/or high costs and (ii) loss of data and potential for technology transfer for the foreseeable future. In either case the effect on both the scientific and broader community will be detrimental.
On the other hand, as the generation of certain data types (e.g. imaging, microarray, phenotypic, etc.) can include costly processes, requiring expensive consumables as well as specialized equipment and personnel for their generation, it can potentially be difficult to fulfil sharing obligations and make resources available, unless there is recognition by peers and investment by funding agencies and the community. This is particularly the case for material resources such as cryopreserved mouse lines, for which rederivation and shipping costs are substantial.
Typically, users want to access data from a single web portal. This can be achieved by storing all the data in one location in a data warehouse but in cases where there are multiple data producers, difficulties with data transfer issues can make a decentralized solution more attractive [8]. Existing technological infrastructures allow the formation of a ‘one-stop-shop’ which brings together data from multiple resources in a single web-interface, enabling collective data querying across different data sets and linking to biological material.
However, in order to achieve such a multi-resource portal, there are several barriers to be overcome in conjunction with some requirements that need to be met. All contributing BRCs should firstly be validated for their data/information quality according to accepted standards, and should be continuously updated, both at the level of material/data as well as incorporation of novel biological resources. To achieve this constantly developing infrastructure, support from both biologists/curators and bioinformaticians is essential, which is a hindrance to the maintenance of a number of these databases. Furthermore, BRCs should all embrace open-access policies upon publication of the related material, or the existence of simple material transfer agreements (MTA) and standards should be implemented so that portals can integrate and become easily interoperable. Such restrictions should be eliminated as much as possible, especially for academic applications, to promote data sharing [11].
Problems encountered
As previously mentioned, one of the biggest concerns that BRCs encounter is their financial sustainability beyond their creation and after the original funding has ended [6]. Typically, BRCs may obtain an initial funding for a project relatively easily where a community need is clear. As a result many biological resource databases have been designed in various research institutes and are commonly created without meeting validated quality standards. Furthermore, they are developed with varying formats and quality, and occasionally exhibit limited international access. Consequently integration of these BRCs into the international data network is often not possible. Searching for mice or ES cells then becomes time consuming and difficult and can result in redundancy of resources. For prolonged data archiving and curation, long term financial support is required which is frequently a stumbling block for BRCs today. Lack of secure funding may frequently result in database or biological resource decommissioning as well as loss of valuable and irreplaceable data. A preferable outcome is the preservation of these unfunded resources by a funded resource with the capacity to do so. The TBASE (http://www.bioscience.org/knockout/knochome.htm) database is an example of this; in this case all the data, which would have otherwise become outdated or lost, was transferred and is now hosted at Mouse Genome Informatics (http://www.informatics.jax.org/) at the Jackson Laboratory. An obvious question that arises is to examine who would provide the required financial support for the archiving of these valuable data and the distribution of biological material, as well as the customer service/user support. How does one support a useful BRC to ensure appropriate data/information archival and curation?
Models examined
Whereas publicly funded BRCs and databases are expected to embrace an open-access policy and be accessible to the broad scientific community, with some notable exceptions, such as the deposition of 300 000 ESTs into GenBank by Merck in 1995, pharmaceutical and biotechnology companies generally do not share data for free. Some companies like Incyte (http://www.incyte.com/), a provider of integrated platforms of genomic technologies, apply a subscription fee, or pay-per-view policy. Other companies, such as Exelixis (http://www.exelixis.com/), employ marketing and public relations policies to help them sell their products or demonstrate their product and technology utility. Finally, some corporations like Wyeth (http://www.wyeth.com/), engage in research collaborations for research they are unable to perform in-house, an effort which indirectly promotes knowledge and dissemination. There are several examples however of very successful public–private models which have, or still do, greatly benefit the not-for-profit sector. For a brief period following their funding crisis in 1996, Swiss-Prot (now Uniprot) a dual-tier system was instituted where for–profits paid an annual fee to the database, whereas academic researchers had free access, effectively being cross-subsidized. Interestingly, Swiss-Prot was ‘re-nationalized’ and completely free access for all users was restored following a large injection of funding from the US NIH in 2002. An example where public–private partnership has successfully collaborated in the long-term is the ‘Structural Genomics Consortium’ (http://www.sgc.ox.ac.uk/) placing protein structures of relevance to human health into the public domain, free from restrictions on use.
Many BRCs currently charge fees to those who want to obtain biological materials and gain access to associated databases. Varying fee structures can be applied for access depending on the nature of the biological material, the status and constraints of the institution holding the resources and its relationship with the public and private sectors, national policies and relevant international frameworks.
There are two major models that have been examined and are currently in use by different BRCs:
Cost recovery
Cost-recovery is defined as recovering the full or partial cost of a project or service, including both its fixed and marginal costs. Typically, it is discussed in the context of cost recovery from users of the services provided, although direct grant funding can be considered as a particular form of cost recovery and is discussed below.
The problems with cost-recovery models for database or, more acutely, BRC sustainability, are articulated by David [12]. In a ‘Ramsey’ model of pricing, the groups that are willing and able to pay more for a fixed level of service over a long period are assigned a larger share of core costs than user groups who are more cost sensitive, and whose inability to pay would be detrimental to the public good. In such a model, the high fixed-cost subscriptions would typically be paid by for-profit organizations, whilst the marginal costs of providing the data or bioresources would be charged to end-users working in universities and not-for-profit organizations. Fixed-cost subscriptions might also be paid by public funding agencies or not-for-profit organizations in respect of their long-term fixed demand for information to allow access to their grantees or employees at a nominal cost. This can be taken to one extreme, as in some of the models for databases discussed below, where one or more agencies cover all fixed and marginal costs by a grant for the common good, and there is no cost passed on to end-users or other organizations. The other extreme is where all costs are recovered from end-users. The problems with the dual layer Ramsey cost-recovery model rest with the willingness of companies and public institutions to fund the high and long-term fixed costs, the segmentation of costs between fixed and distributive components (not an easy calculation to make given the complex ongoing activities of open biological databases) and what would be considered a fair cost for the service to individual users, whilst still maintaining public good. We are not aware of any resource that has been able to recover all costs through a single layer system with fixed and marginal costs recouped directly from individual end-users. David also argues against such an approach, especially for databases in which the true marginal cost of each transaction is actually very low: ‘commitment to implement them (user charges) on the part of the rich societies would most likely result in pricing the use of scientific information and data beyond the reach of many poorer societies’. There is a political and social dimension here, along with the general issue of the potential harm to the rate of scientific advance caused by the imposition of cost barriers to data and materials access.
However, a partial ‘cost-recovery’ model, where marginal and a variable fraction of fixed costs are recouped from end-users, is a clearly viable funding approach for the partial support of core services which augments direct grant funding to the resource; a useful strategy used for income supplementation in order to sustain the running of infrastructure. In some current examples, core costs are met by one funding agency for the benefit of all, for example the MMRRCs in the USA where core funding is provided by the National Institutes of Health (NIH) [13], and some of the marginal cost recovered from end-users, or as with the Human Genome Variation database (HGV-G2P; http://www.hgvbaseg2p.org/index) where there are multiple agencies and organizations covering both the core and marginal costs and data is freely available [14].
In the case of the BRCs, a cost-recovery model by which revenue might be secured to support the infrastructure is ‘fee-for-service’, but despite the contribution of fees we are not aware that even true full marginal costs are met in this way by any BRC, let alone core costs, and income from ‘fee-for-service’ has a significant but only mitigating effect on the overall cost of running the core resource. This is a consequence of setting the marginal costs, in the Ramsey sense, at levels which are sustainable for investigators on fixed grant income, and to a large degree tacitly negotiated with funding agencies as to what level of cost is acceptable.
Several BRCs utilize the partial cost-recovery model in conjunction with other methods of financial support. One example of this is the EMMA which provides a free archiving repository for mouse mutant lines. The bulk of the costs of actually distributing the mice are expected to be recovered from the fees charged to both for- and not-for-profit end-users who order the material. However, the considerable costs associated with archiving the mice, supporting the informatics and project organization are still provided from a European Commission FP7 grant as well as national research programmes and institutional funds at individual partner sites. Other examples include the Drosophila Genomics Resource Center (https://dgrc.cgb.indiana.edu/), the Bloomington Drosophila Stock Center at Indiana University (http://fly.bio.indiana.edu/) and the John Innes Centre Genome Laboratory (JGL; http://jicgenomelab.co.uk/).
Institutional funding
A common model for the financial sustainability of a resource is through allocated funds obtained from a single public institution towards the respective BRC. This approach is most commonly applied to data resources. An example of this model are the databases operated by the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) which receives funds from both the National Library of Medicine (NLM; http://www.nlm.nih.gov/) and the NIH (http://www.nih.gov/).
The role of industry versus the role of government
Both industry and government agencies have provided support to BRCs. Some BRCs use a dual support system, where public research agencies and not-for-profit organizations provide grants for specific projects and programs, which may involve consortia of laboratories, whereas government funding agencies provide block grant funding to Research Institutes to support the research infrastructure and enable the institutions to undertake ground-breaking research of their choosing. Such funding also provides the capacity to undertake research commissioned by the private sector, government departments, charities, the European Union and other international bodies. The European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/) is a good example of this dual funding support practice, as it is funded by the governments of EMBL's member states, as well as other major funders such as the European Commission, Wellcome Trust, US National Institutes of Health, UK research councils and some industry partners.
Furthermore, there are specific ‘projects’ (e.g. biobanks—collections of cells, tissues, blood, DNA samples) that may have a two-fold character, as collections of both samples and data. These may be operated under the auspices of either the public sector institutions (i.e. university departments) or of individual or private bodies (e.g. pharmaceutical companies). Irrespective of the responsible institution, they may be funded from public or private resources. One could expect that some funding from these projects may be dedicated towards resource integration and dissemination. Good examples of this are mutant cryorepositories which provide the facility to cryopreserve and distribute mouse lines as sperm or frozen embryos. These are typically partly institutionally funded but receive additional funding from specific projects, such as the EU-funded EMMA project.
Development of the business model aforementioned, as a supplementary activity towards cost recovery, is not as effective for underpinning the infrastructure, as it does not cover Full Economic Costing (real costs of running an infrastructure, including all costs above and beyond consumables and direct staff costs; these involve rent for space, overheads, staff salary/benefits, staff training and any business development support) or opportunity costs.
With regard to industry investment, association through advertising could be seen as a potential revenue generator, useful to provide valuable support towards further developing the resource assets (e.g. validated assays, new applications). However, it is doubtful that the benefits would be enough to cover the infrastructure and the business development overhead will outweigh any overall benefit. In order for companies to be drawn towards website advertising, visibility of at least 10 000 visitors per month is required [5]. Considering the specialization of biological databases, even many of the big ones do not have this volume of traffic, and therefore attracting commercial clients makes it almost impossible to raise enough revenue to support BRCs.
A model with potential: academic–commercial partnership on core competencies
Another model that has been examined and appears to have great potential in being successful towards the prolonged financial sustainability of BRCs is an ‘academic-commercial partnership’. Academic laboratories, mostly sustained by institutional funding, or grants, develop new applications and tools as well as analysis systems, whereas concurrently they support the identification of communal needs and define quality standards all of which prove to be beneficial to the research community. Commercial organizations on the other hand, which are financially supported by their own commercial activities, function in a collaborative way between research and licensing (Pharmaceutical and Biotechnology companies) and operate as service providers, offering standard technologies and quality systems, sales and marketing distributors.
In the context of CASIMIR, and in the course of examining the potential financial models that resource centres could adopt for their maintenance, the MUGEN Mouse database (MMdb; www.mugen-noe.org/database/), a virtual mutant mouse repository created in the context of the MUGEN Network of Excellence (www.mugen-noe.org/) to provide on-line information on murine models for immunological disease [15], serves as a use-case example. For demonstration purposes, MMdb, taking advantage of its simplicity and useful size, currently provides direct trial links, under the gene information, to Invitrogen (http://www.invitrogen.com/) and Geneservice (http://www.geneservice.co.uk/) through the gene IDs (Figure 1). The user may therefore be directly transferred to the respective product page, where all the gene-related products (e.g. antibodies, RNAi, primers, cDNA clones, proteins, assay kits, etc.) are presented. Ensuing the overall discussions regarding financial sustainability of databases, and following a successful connection, MMdb has approached Invitrogen as well as other potential companies, asking them to link their individual products with the respective mouse model and also examine the possibility that such big vendor corporations would be interested in linking with MMdb and explore their willingness towards marketing/advertisement service charges which could help maintain the database. Indeed Invitrogen responded very positively towards this effort, and has pledged to undertake a survey with regard to the company's perspectives and willingness to financially support this effort. Unfortunately, the overall response was not as expected, since only one out of the six companies approached responded to the request, demonstrating some enthusiasm and feedback in this attempt. The suggested approach, although in principle appearing to have great potential, in practice is somewhat harder to achieve, as companies are not that willing to sponsor academic institutions. This may of course be a matter of time and should big vendor corporations be appropriately primed this arrangement may indeed prove to be beneficial towards prolonged sustainability. Finally, such an approach would only be applicable provided the fundamental unit of information is related to company goods (e.g. reagents) and will therefore only apply to a fraction of databases.
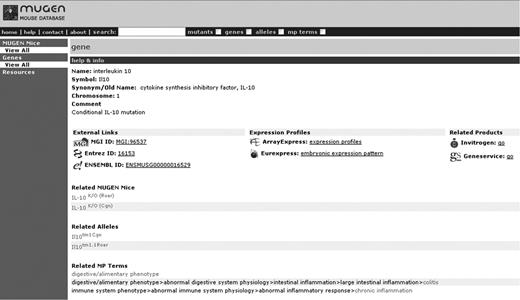
Sample screen shot of MMdb ‘IL-10’ gene with the direct trial links, under the gene information, to Invitrogen and Geneservice through the gene ID.
The role of consortia
The European Commission in support of the fifth and sixth Framework Programmes has over the last seven years sponsored a number of projects generating biological experimental data, including sequences, and material resources such as biological collections. Some of these consortia (e.g. EUMORPHIA, EUCOMM, EUMODIC, EUREXPRESS, EMMA, MUGEN, etc.) also serve as liaisons towards the European Commission, giving advice with respect to specific areas of interest and their respective needs for further development and also suggesting potential future directions that the European Commission should pursue.
Furthermore, the European Commission has also supported some co-ordination actions (e.g. PRIME, CASIMIR) especially to organize and bring together the individual European efforts as well as survey the scientific community needs. These consortia also play an intermediary role between the scientific community and the European Commission, making recommendations to the latter with respect to the needs that the scientific community has, thus aiming to improve scientific development. This interactive relationship allows networks to lobby both national and international funders, for example, to improve application practices and for funders to approach and consult with the network with regard to issues and priorities.
The CASIMIR consortium, in an effort to evaluate the financial sustainability models adopted by existing biological databases and resources, has examined the responses obtained from MRB's (Mouse Resource Browser) online questionnaire (http://bioit.fleming.gr/imouse/). The overall response was satisfactory; indeed there were 79 responses obtained covering resources from around the world. The majority of responders were of European origin (51%), followed by American resources (38%), while the remaining 11% of obtained responses were from databases and resources from Canada, Japan and Australia (Figure 2). As expected, the majority of biological resources and databases were created as part of a particular funded project. Of the 50 resources that responded to this particular question, 68% confirmed that their database or resource indeed originated from a funded project, while the remaining 32% did not (Figure 3). On the other hand, despite the fact that in the majority of cases the original financial support came from funded projects, upon completion/expiration of these three- or five-year projects, financial maintenance of the respective resource was achieved through Institutional or Government funding (44 and 36%, respectively), while only the remaining 20% of biological databases were funded by other sources such as the industry (4%) (Figure 4). Through examination of the financial maintenance achieved by some resource centers for sustaining their core activities and assessment of how these may be applied towards long term preservation of databases it has become evident that most databases obtain an initial funding for a particular project, and then need to be further maintained through institutional or government funds.
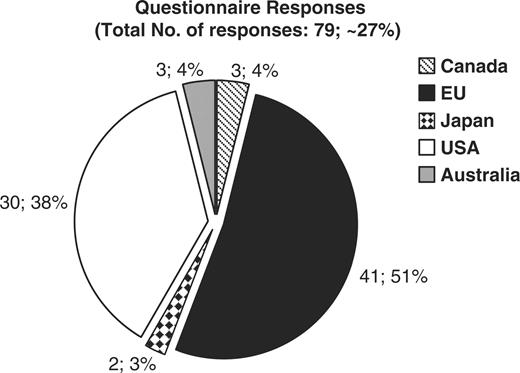
Graph representing the origin of each biological database or resource that responded to the online questionnaire. 51 percent of resources are in Europe, 38% in the USA, 4% in Australia and Canada, and 3% in Japan.
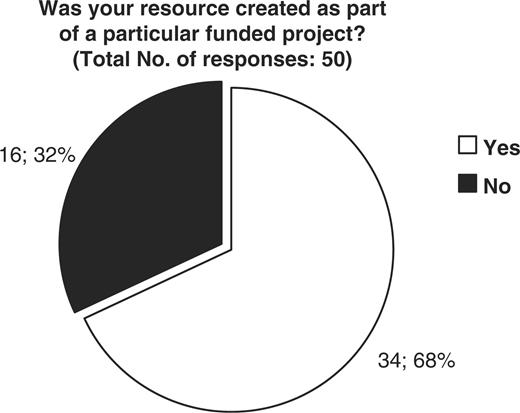
Representation of the financial support originally obtained for the creation of each biological database or resource. 68 percent of resources were created as part of a particular funded project, while 32% were not.
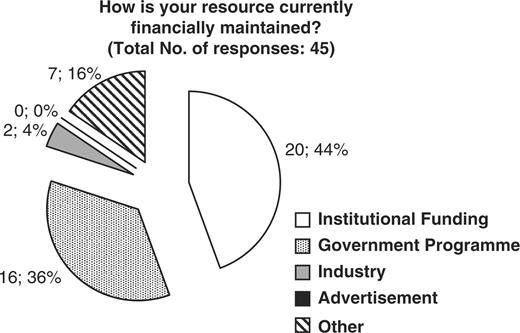
Representation of the financial support currently provided to maintain biological databases and resources. 44 percent of resources subsidize through institutional funds, 36% through Government programs, 4% from Industrial funds and 16% from other sources.
Recommendations for the mouse functional genomics community
Having reviewed extensively the substantial amount of information provided by BRCs and the importance of making the data freely available to the research community, it is clear that it is imperative to promote data preservation and dissemination, for secure storage and easy retrieval of information. Moreover, it will often be appropriate that BRC databases should not exist as classical data warehouses, but rather a cluster of activities supporting the community of academic and commercial researchers all aiming, through a unified effort, towards providing information for the progression of research. CASIMIR is indeed already taking action in the direction of promoting database integration and interoperability, and should investigators conform to their responsibilities and share data as recommended by UPSIDE [7,8] this would obviously greatly promote research activity.
Furthermore, following the close examination of setbacks that most of these BRCs today encounter and existing business models that they could potentially adopt in order to reinforce database sustainability, the conclusion that can be drawn is that long-term sustainability of databases requires adequate and reliable sources of funding so that data is preserved and disseminated properly.
With regard to the business models examined in this manuscript as potential patterns to be adopted by BRCs for their financial sustainability, the ‘full cost recovery’ model which has already been tested by some resources has proved to not be viable for data resources. The ‘fee-for-service’ or ‘partial cost-recovery’ model is already practiced, at least in part, by some BRCs. For data provided this is contrary to the UPSIDE recommendations, according to which data should be shared, but in practice most BRCs employing this approach are providing material resources, which have substantially higher costs and it is open to debate if these can reasonably be provided completely free of charge. The most promising model examined in this manuscript is ‘Institutional Funding’ which seems to provide a secure environment for the BRCs to develop and implement a secure data management plan and potentially ensure the long-term accessibility of the related project data. Indeed agencies around the world such as the NIH and the EU through ELIXIR (http://www.elixir-europe.org), are now turning their attention to working out how best to assist the growth of validated and accessible databases. This should involve, at the least, development of policies for evaluating proposals on databases and associated analytic tools, for their sustained funding, and for ensuring that the data deposited remain accessible long after the project originators have moved on. The aforementioned model of academic-commercial partnership may appear to have potential should vendor corporations become involved in this collaborative effort. In all cases, funders should be aware of the need to support viable career paths for the software engineers and bioinformaticians who create the knowledge environments and curate the data in them. In order to obtain value for money, it will be vital for funding agencies to carefully select the databases they choose to support and then to support them for the long term. They must encourage the sustained availability of these data and build incentives for the development of cross-querying capability.
Discussion
The last decade has seen a rapid growth in the genome sciences, through modern advances in biological sciences, molecular biology and genetics, which have enabled genome-wide analysis in most model organisms, and the generation of high-throughput of data. To facilitate the secure storage and easy retrieval of this substantial amount of information, numerous data and biological material resources have been created which are of significant value and should be openly accessible to all scientists for the purposes of result validation, testing new hypotheses and developing new technologies/platforms. An inevitable consequence that has arisen from this data and biological material resource boom is the significant challenge in the access and sustainability of these databases. Preservation of these centralized repositories is therefore imperative. CASIMIR continues to review the potential business models that biological resources could adopt for their financial sustainability and prolonged data storage and aims to appropriately make recommendations to the funding agencies and the community at large.
Funding
Sixth Research Framework Programme of the Commission of the European Community (CASIMIR LSH-2005-1.1.0-1, MUGEN LSHG-CT-2005-005203). Funding for open access charges: CASIMIR.
Conflict of interest statement. None declared.
Acknowledgements
The authors would like to thank all CASIMIR members as well as participants to the various CASIMIR meetings for fruitful discussions.



