-
PDF
- Split View
-
Views
-
Cite
Cite
P. Gaudet, L. Lane, P. Fey, A. Bridge, S. Poux, A. Auchincloss, K. Axelsen, S. Braconi Quintaje, E. Boutet, P. Brown, E. Coudert, R.S. Datta, W.C. de Lima, T. de Oliveira Lima, S. Duvaud, N. Farriol-Mathis, S. Ferro Rojas, M. Feuermann, A. Gateau, U. Hinz, C. Hulo, J. James, S. Jimenez, F. Jungo, G. Keller, P. Lemercier, D. Lieberherr, M. Moinat, A. Nikolskaya, I. Pedruzzi, C. Rivoire, B. Roechert, M. Schneider, E. Stanley, M. Tognolli, K. Sjölander, L. Bougueleret, R.L. Chisholm, A. Bairoch, Collaborative annotation of genes and proteins between UniProtKB/Swiss-Prot and dictyBase, Database, Volume 2009, 2009, bap016, https://doi.org/10.1093/database/bap016
Close - Share Icon Share
Abstract
UniProtKB/Swiss-Prot, a curated protein database, and dictyBase, the Model Organism Database for Dictyostelium discoideum, have established a collaboration to improve data sharing. One of the major steps in this effort was the ‘Dicty annotation marathon’, a week-long exercise with 30 annotators aimed at achieving a major increase in the number of D. discoideum proteins represented in UniProtKB/Swiss-Prot. The marathon led to the annotation of over 1000 D. discoideum proteins in UniProtKB/Swiss-Prot. Concomitantly, there were a large number of updates in dictyBase concerning gene symbols, protein names and gene models. This exercise demonstrates how UniProtKB/Swiss-Prot can work in very close cooperation with model organism databases and how the annotation of proteins can be accelerated through those collaborations.
Introduction
Dictyostelium discoideum is a unicellular eukaryote often referred to as a social amoeba because it can form a multicellular structure when nutrient conditions are limiting. It is a powerful model system for the study of signal transduction, cell migration, the cytoskeleton, developmental morphogenesis and secondary metabolism (1). D. discoideum belongs to the supergroup Amoebazoa, division Mycetozoa (2). Phylogenetic analysis places Dictyostelium in one of the earliest branches to emerge after the divergence of plants and animals. Dictyostelium has retained more of the diversity of the ancestral eukaryotic genome than have plants, animals or fungi and is one of the best-studied organisms in that central phylogenetic position. The Dictyostelium genome sequence was published in 2005 and contains ∼12 500 protein-coding genes [(3), Table 1].
| Genome size | 34 Mbp |
| Chromosomes | 6 |
| Protein coding genes | ∼12 500 |
| Genes with splice variants | ∼20 identified |
| Curated pseudogenes | 160 |
| Repetitive elements | 500–1000, clustered |
| rRNA genes | 8 (100 copies, extrachromosomal) |
| tRNA genes | 418 |
| Other non-coding RNAs | ∼100 |
| Genome size | 34 Mbp |
| Chromosomes | 6 |
| Protein coding genes | ∼12 500 |
| Genes with splice variants | ∼20 identified |
| Curated pseudogenes | 160 |
| Repetitive elements | 500–1000, clustered |
| rRNA genes | 8 (100 copies, extrachromosomal) |
| tRNA genes | 418 |
| Other non-coding RNAs | ∼100 |
| Genome size | 34 Mbp |
| Chromosomes | 6 |
| Protein coding genes | ∼12 500 |
| Genes with splice variants | ∼20 identified |
| Curated pseudogenes | 160 |
| Repetitive elements | 500–1000, clustered |
| rRNA genes | 8 (100 copies, extrachromosomal) |
| tRNA genes | 418 |
| Other non-coding RNAs | ∼100 |
| Genome size | 34 Mbp |
| Chromosomes | 6 |
| Protein coding genes | ∼12 500 |
| Genes with splice variants | ∼20 identified |
| Curated pseudogenes | 160 |
| Repetitive elements | 500–1000, clustered |
| rRNA genes | 8 (100 copies, extrachromosomal) |
| tRNA genes | 418 |
| Other non-coding RNAs | ∼100 |
UniProtKB/Swiss-Prot (http://www.uniprot.org/) is the cornerstone of UniProtKB, the comprehensive resource developed by the Universal Protein Consortium (4). UniProtKB/Swiss-Prot is a manually curated knowledgebase that provides high-level annotation of proteins, including the description of the function and catalytic activity, domain structure, post-translational modifications and variants. One of its goals is to be highly integrated with other databases providing complementary data.
dictyBase (http://www.dictybase.org) is the model organism database for D. discoideum. dictyBase annotators curate gene models as well as the literature relevant to Dictyostelium. Gene predictions obtained from the Dictyostelium sequencing project are manually reviewed by curators. The evidence for the gene model, including ESTs, sequence similarity and start, stop and intron boundaries are analyzed before the gene model is approved ‘as is’ or modified by the curator using Apollo (5). Approximately 15% of the gene models require some modification. Moreover, since the automated tools do not detect splice variants, those are added manually. Less than 0.5% of Dictyostelium genes have evidence for splice variants. In addition to curating gene models, dictyBase curators annotate a gene product's function, and Gene Ontology terms. This information is provided from the literature when available, or based on sequence similarity. Finally, strains and phenotypes are curated from publications.
As just described, much of the data captured by the two databases is complementary. Of the 12 500 proteins encoded by the Dictyostelium genome, only 537 were annotated in UniProtKB/Swiss-Prot in January 2008, and most of those entries were done before the Dictyostelium genome sequence became available. The remaining 12 000 sequences—automatically translated from the genome and awaiting manual curation—were made available in UniProtKB/TrEMBL, the automatically generated part of UniProtKB. dictyBase was created in 2003 and, as of January 2008, had annotations for nearly 5000 genes (6). The large discrepancy in the extent of manually curated data relevant to Dictyostelium between the two resources prompted us to find a method to perform annotations as efficiently as possible by making maximal use of the information available from both resources. This was achieved by a collaborative ‘annotation marathon’, where curators from both groups worked together to annotate over 1000 genes and proteins in the course of one week.
Results and discussion
Goals of the marathon
Before this collaboration, there was no annotation program at UniProtKB/Swiss-Prot focused on Dictyostelium, which means that few entries were annotated and for the most part those were out of date. The main objective of the marathon was to improve the curation of Dictyostelium UniProtKB/Swiss-Prot entrie jump start.
The week-long D. discoideum protein annotation marathon organized jointly by the SIB Swiss-Prot group of the UniProt consortium and dictyBase took place in Geneva in March 2008. A total of 28 UniProt annotators and two dictyBase curators participated in the marathon. Over the full week this represented 23 full-time employees for annotation and approximately three people for correction and integration of the annotated entries.
Annotation targets
Two sets of entries were the object of the annotation marathon: First, entries that correspond to conserved eukaryotic proteins, which can relatively easily be annotated based on sequence similarity. These entries were selected by Kimmen Sjölander's group at UC Berkeley. Using pre-computed phylogenetic trees in the PhyloFacts phylogenomic encyclopedias (7), they extracted a set of about 1500 Dictyostelium proteins that have orthologs in at least two other major eukaryotic phyla (human/mouse and/or Drosophila and/or S. cerevisiae/S. pombe; the complete list is provided in supplemental file 1) using the PHOG orthology prediction algorithm (8). The second set of target entries were those for which there is published literature. Annotators were asked to use information already present in dictyBase and to extract more data from the selected articles as quickly as possible. About 100 articles were distributed (an average of three per annotator), covering ∼50 genes. To accelerate the process, proteins with a large body of literature were excluded. Since some proteins from the PhyloFacts analysis were also linked to publications, a total of 284 articles were read during the marathon. The annotation statistics resulting from the marathon are summarized in Table 2. During the five days of the marathon, 1044 UniProtKB/Swiss-Prot entries were annotated. Modifications done at UniProtKB/Swiss-Prot can be viewed by clicking the ‘History’ link on each page; for example, the changes made during the marathon for the Q54U87 record can be viewed by comparing annotation versions 28 and 29 of the dhkA entry, here: http://www.uniprot.org/uniprot/Q54U87?version=28&version=29. This gene was previously not reviewed in UniProtKB/Swiss-Prot; during the marathon, it was integrated into UniProtKB/Swiss-Prot, and annotated with the gene name, protein function, E.C. number, relevant articles, protein functional domains, etc. The list of entries annotated during the marathon is provided in Supplementary Data 2.
| Number of proteins annotated during the 5 days of the actual annotation marathon | 1044 |
| Total number of papers read/annotated | 284 |
| Gene names changed/added in dictyBase | 254 |
| Name changes still being processed | 88, including 40 with naming disagreements |
| Gene models corrected in dictyBase | 27 |
| Number of proteins annotated during the 5 days of the actual annotation marathon | 1044 |
| Total number of papers read/annotated | 284 |
| Gene names changed/added in dictyBase | 254 |
| Name changes still being processed | 88, including 40 with naming disagreements |
| Gene models corrected in dictyBase | 27 |
| Number of proteins annotated during the 5 days of the actual annotation marathon | 1044 |
| Total number of papers read/annotated | 284 |
| Gene names changed/added in dictyBase | 254 |
| Name changes still being processed | 88, including 40 with naming disagreements |
| Gene models corrected in dictyBase | 27 |
| Number of proteins annotated during the 5 days of the actual annotation marathon | 1044 |
| Total number of papers read/annotated | 284 |
| Gene names changed/added in dictyBase | 254 |
| Name changes still being processed | 88, including 40 with naming disagreements |
| Gene models corrected in dictyBase | 27 |
There were 27 genes whose models were modified at dictyBase following suggestions by UniProtKB/Swiss-Prot annotators. One such modified gene model is commd10 (DDB_G0275249): the gene prediction used an incorrect splice site. The correct splice donor, located 21 nucleotides downstream, is supported by sequence similarity (Figure 1).
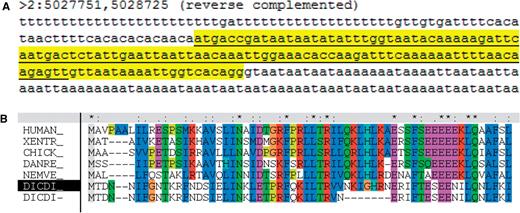
A modified gene model, commd10 (DDB_G0275249). (A) The gene prediction (underlined) selected a splice donor (gttaat) 21 nt upstream from that (gtaata) of the curated gene model (highlighted in yellow). This image was generated using the dictyBase Genome Browser. (B) The modified gene prediction is supported by sequence similarity. The new Dictyostelium gene model is indicated by a black shading of ‘DICTY’ on the labels on the left; the new gene model produces a better alignment than the gene prediction (bottom row), indicated by the gap in the alignment of the latter.
Improvements to UniProtKB/Swiss-Prot annotations
UniProtKB/Swiss-Prot always tries to provide uniform annotation across families, orthologs as well as paralogs. During the Dictyostelium annotation marathon, 60 protein families were created, in which 249 new entries from various organisms, including fungi, plants and mammals were included. There were 115 non-Dictyostelium UniProtKB/Swiss-Prot entries that were updated, some at the level of the sequence itself, others for protein names, or for information relative to domains, ligands, post-translational modifications, etc.
Nomenclature issues
Gene nomenclature is a difficult item to standardize and caused some of the most difficult discussions between dictyBase and UniProtKB/Swiss-Prot annotators, some of which are still unresolved.
Dictyostelium gene names are based on the Demerec system (three small case letters followed by a capital letter if there is more than one member of the gene group). The names are derived either from protein function or from a phenotype, for example, acaA for adenylate cyclase and sadA for substrate-adhesion mutant. In the absence of published names, dictyBase curators assign names based on sequence similarity; hence gene names do not always conform to Demerec nomenclature. A number of genes cannot be named based on those rules and retain the dictyBase gene ID (starting with DDB_G) as their primary name.
The work flow at UniProtKB/Swiss-Prot make it easy to assign the same name to homologous genes from several different species at once. This is very helpful for cross-species comparisons, but is not always possible: many genes have been published under certain names and cannot be changed without creating confusion. Large gene families with lineage-specific duplications pose another difficulty, as using the same name across species might mislead users concerning homologous and orthologous relationships between the different genes. Yet another problem concerns names that imply the presence of other proteins, such as ‘p53-interacting protein’, which is meaningless for an organism lacking p53, or names that refer to diseasens or organs absent from the organism being annotated. Two-hundred and twenty gene names were added or changed in dictyBase. Having Dictyostelium as one of the species annotated at UniProtKB/Swiss-Prot helps ensure that names proposed for propagation are relevant for a more comprehensive taxonomic coverage.
Integrating protein information in dictyBase
As mentioned earlier, dictyBase aims to annotate all information relevant to the Dictyostelium genome. UniProtKB/Swiss-Prot annotates exclusively proteins and has more precise protein annotations than dictyBase does. Instead of reproducing the work done by expert protein curators at UniProtKB/Swiss-Prot, dictyBase has created a novel ‘Protein Page’ that displays the protein information that used to be presented in the dictyBase ‘Gene Page’, such as gene product name, protein length, and molecular weight. In addition, this page now includes information from UniProtKB/Swiss-Prot that is not specifically annotated by dictyBase, such as sequence processing, sequence existence evidence, subunit structure, post-translational modifications, as well as sub-cellular location. An example is shown in Figure 2 for the mhcA gene (DDB_G0286355). Although this gene is fully annotated with respect to the information captured by dictyBase, extra information concerning the protein product can be obtained from UniProt/KB/Swiss-Prot. This data is updated biweekly.
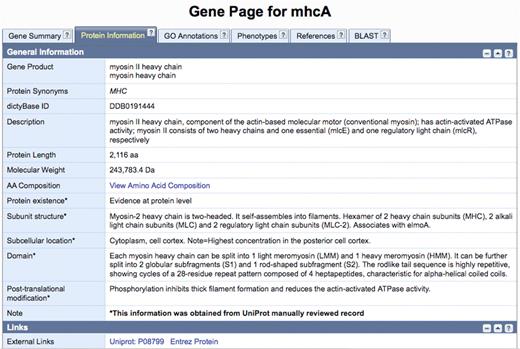
The new dictyBase protein information page will display annotations from UniProtKB/Swiss-Prot. Information for the MhcA protein (gene ID: DDB_G0286355; sequence ID: DDB0191444) parsed from UniProtKB/Swiss-Prot record P08799 includes sequence processing, sequence existence evidence, subunit structure, post-translational modifications, as well as sub-cellular location. Fields obtained from UniProtKB/Swiss-Prot are marked with an asterisk.
The continuation of the UniProtKB/Swiss-Prot—dictyBase collaboration
The numerous entries modified during the annotation marathon raised several annotation issues that were not all solved during the marathon week. For example, several gene models were dubbed questionable (as described in Figure 1) and have been submitted to dictyBase curators for review, but not all could be fixed during the marathon week but were adjusted since. Also, a number of gene nomenclature suggestions from Swiss-Prot curators were in conflict with dictyBase guidelines and were discussed and adjusted during and after the marathon.
The momentum generated by the annotation marathon prompted us to continue working together with the long-term goal to have Dictyostelium completely annotated in Swiss-Prot. The goal of annotating the complete Dictyostelium proteome is facilitated by the high quality of the genome and annotations at dictyBase. Two Swiss-Prot curators continue to spend a significant portion of their time annotating Dictyostelium entries, while a few others contribute more occasionally, for an average of roughly 2.2 FTEs per year. Those have annotated ∼1800 entries in the year following the marathon. The following priority targets represent low-hanging fruits that can lead to annotated entries with minimal effort:
(a) Orphan entries: these are proteins that have no similarities outside of Dictyostelium. Those were identified by BLAST analysis of all the Dictyostelium proteins lacking InterPro hits against UniProtKB. From those analyses, 1034 proteins specific to Dictyostelium (“ORFans”) were identified. The UniProt Anabelle sequence analysis platform was then used in batch mode to semi-automatically annotate these entries. This software platform can predict sequence features such as transmembrane regions, signal or transit peptides as well as coiled-coil motifs that can be pertinent to ORFan annotation. This approach provided many entries easy to annotate; this group of proteins also contained a large fraction of genes derived form retrotransposons, pseudogenes, and incorrect gene predictions (thus their ‘orphan’ status) and several gene models ended up being deleted.
(b) Large protein families that have already been annotated by dictyBase or that are scheduled to be annotated by them. Annotation of the mitochondrial carrier proteins, cytochromes P450, polyketide synthases, hssA/B family members, and the GATA and the Myb family transcription factors have already been carried out. The next targets will be the ABC transporters, acetyl-CoA synthetases, bZIP transcription factors, HOX transcription factors, short chain dehydrogenases, protein kinases, ribosomal proteins, vacuolar protein sorting, small GTPases, GEFs and GAPs.
(c) Specific targeting of proteins that are linked with publications that have already been curated in dictyBase with as much information transfer as possible directly from dictyBase (either as Gene Ontology (GO) terms or in the “Summary” section).
The result of this sustained annotation effort is highlighted in Table 3: the number of Dictyostelium entries has been multiplied by 10 in two years, and Dictyostelium is now the 10th most represented organism in the manually curated UniProtKB/Swiss-Prot database. Annotation is a never-ending process and must be updated as new data becomes available. The extensive collaboration between the UniProt consortium and dictyBase makes it easier to flag entries that need to be annotated or updated.
| Release . | Date . | Number of Dicty entries . | Rank in Swiss-Prot . |
|---|---|---|---|
| 51.4 | January 2007 | 337 | 165th rank |
| 55.0 | February 2008 | 537 | 86th rank |
| 55.2 | April 2008 | 1803 | 18th rank |
| 57.1 | April 2009 | 3619 | 10th rank |
| Release . | Date . | Number of Dicty entries . | Rank in Swiss-Prot . |
|---|---|---|---|
| 51.4 | January 2007 | 337 | 165th rank |
| 55.0 | February 2008 | 537 | 86th rank |
| 55.2 | April 2008 | 1803 | 18th rank |
| 57.1 | April 2009 | 3619 | 10th rank |
| Release . | Date . | Number of Dicty entries . | Rank in Swiss-Prot . |
|---|---|---|---|
| 51.4 | January 2007 | 337 | 165th rank |
| 55.0 | February 2008 | 537 | 86th rank |
| 55.2 | April 2008 | 1803 | 18th rank |
| 57.1 | April 2009 | 3619 | 10th rank |
| Release . | Date . | Number of Dicty entries . | Rank in Swiss-Prot . |
|---|---|---|---|
| 51.4 | January 2007 | 337 | 165th rank |
| 55.0 | February 2008 | 537 | 86th rank |
| 55.2 | April 2008 | 1803 | 18th rank |
| 57.1 | April 2009 | 3619 | 10th rank |
Producing eukaryotic-wide automatic annotation rules for UniProtKB
A large number of Dictyostelium proteins annotated in UniProtKB/Swiss-Prot belong to conserved families covering a wide taxonomic range and generally having a single copy in each genome (few in-paralogs and between species paralogs). Therefore it is both feasible and desirable to use Dictyostelium as a ‘seed’ organism for an approach that would quickly produce eukaryotic-wide templates designed to automate the annotation of orthologs. Those templates consist of all annotations from a group of entries from homologous proteins that can confidently be propagated across species. This approach is an extension to that already successfully used by the UniProtKB/Swiss-Prot HAMAP project (9) in the context of the automatic annotation of proteins from complete prokaryotic and archaeal proteomes.
We currently have a set of 1320 Dictyostelium candidate entries that are sufficiently conserved to be used to contribute to those annotation templates, and we expect this number to continue to grow. The templates will be created as follows. A BLAST analysis of the candidates will be performed against a subset of UniProtKB composed of complete eukaryotic proteome sets that includes sequences from mammals, fungi, insects, nematodes, plants, anemone, hydra, etc, in order to eliminate potential problematic cases such as those that underwent a significant lineage-specific expansion in some of the available phyla from the starting sets. Multiple alignments will be automatically created for all seed candidates and then manually checked in order to exclude or correct erroneous gene model predictions. To create the annotation templates, the entries of the seed candidates, including the Dictyostelium entries, will be checked to make sure that they are species-neutral and that proposed recommended protein and gene names are correct and up to date. The annotation templates will be automatically applied to un-annotated UniProtKB/TrEMBL entries and, following manual quality controls, be entered into UniProtKB/Swiss-Prot.
Propagation of annotations across species is a complex endeavor and the UniProtKB/Swiss-Prot team strives to be as conservative as possible in what type of annotations can safely be transferred. After nine years of building and running the HAMAP pipeline, the results of this large scale experiment warrant that the UniProt consortium embark in a similar project targeting eukaryotic organisms. This is a long-term project which will be the subject of further publications.
Conclusions and perspectives
This marathon and the resulting ongoing collaboration is extremely positive for both UniProtKB/Swiss-Prot and dictyBase. We believe such collaborative efforts are invaluable for the contributing databases and by extension to all of their users. We hope this will encourage other groups to undertake similar collaborations.
Funding
National Institutes of Health to dictyBase [GM64426, HG0022]. UniProt is mainly supported by the National Institutes of Health grant (2U01HG02712-04). UniProtKB/Swiss-Prot activities at the SIB are supported by the Swiss Federal Government through the Federal Office of Education and Science. Funding for open access charge: NSF grant no. 0732065, The PhyloFacts Phylogenomic Encyclopedia of Microbial Protein Families.
Conflict of interest statement. None declared.
Acknowledgements
The authors thank Pierre Cosson and Thierry Soldati for providing to the Swiss-Prot annotators a very good overview of the biology of Dictyostelium and for their availability during the annotation marathon to answer questions.



