-
PDF
- Split View
-
Views
-
Cite
Cite
Neil D. Rawlings, A large and accurate collection of peptidase cleavages in the MEROPS database, Database, Volume 2009, 2009, bap015, https://doi.org/10.1093/database/bap015
Close - Share Icon Share
Abstract
Peptidases are enzymes that hydrolyse peptide bonds in proteins and peptides. Peptidases are important in pathological conditions such as Alzheimer's disease, tumour and parasite invasion, and for processing viral polyproteins. The MEROPS database is an Internet resource containing information on peptidases, their substrates and inhibitors. The database now includes details of cleavage positions in substrates, both physiological and non-physiological, natural and synthetic. There are 39 118 cleavages in the collection; including 34 606 from a total of 10 513 different proteins and 2677 cleavages in synthetic substrates. The number of cleavages designated as ‘physiological’ is 13 307. The data are derived from 6095 publications. At least one substrate cleavage is known for 45% of the 2415 different peptidases recognized in the MEROPS database. The website now has three new displays: two showing peptidase specificity as a logo and a frequency matrix, the third showing a dynamically generated alignment between each protein substrate and its most closely related homologues. Many of the proteins described in the literature as peptidase substrates have been studied only in vitro. On the assumption that a physiologically relevant cleavage site would be conserved between species, the conservation of every site in terms of peptidase preference has been examined and a number have been identified that are not conserved. There are a number of cogent reasons why a site might not be conserved. Each poorly conserved site has been examined and a reason postulated. Some sites are identified that are very poorly conserved where cleavage is more likely to be fortuitous than of physiological relevance. This data-set is freely available via the Internet and is a useful training set for algorithms to predict substrates for peptidases and cleavage positions within those substrates. The data may also be useful for the design of inhibitors and for engineering novel specificities into peptidases.
Database URL:http://merops.sanger.ac.uk
Introduction
Peptidases (proteases or proteinases) are enzymes that hydrolyse the peptide bonds between amino acids in a protein or peptide chain. Hydrolysis of such bonds is required for removal of targeting signals (signal and transit peptides (1), ubiquitin (2), SUMO (3) and NEDD8 (4) peptides), the release of a mature protein from its precursor (5), the switching off of a biological signal by degradation of the signal protein (6), and for widespread catabolism of proteins for recycling of the amino acids. When proteolysis occurs unchecked, then diseases can result, such as Alzheimer's (7), osteoarthritis (8), emphysema (9), tumour invasion (10) and acute pancreatitis (11). Pathogens use peptidases to enter the host and to degrade host proteins for food (12).
Peptides and proteins have been widely used to characterize the specificity of peptidases, but frequently the substrates chosen have been physiologically irrelevant. One of the most popular substrates has been the oxidized insulin B-chain, because this is a peptide without tertiary structure, and cleavage depends solely on the preference of the peptidase (13). (The terms ‘specificity’, ‘selectivity’ and ‘preference’ are used interchangeably here.) However, peptidase preference is exactly that: a preference only. Researchers often find that after prolonged exposure to a peptidase other bonds are degraded, albeit slowly, once none of the preferred bonds remain. If the peptidase preparation is not pure, then there is the danger that some of the observed cleavages are due to contaminating peptidases.
The bond in a substrate where hydrolysis occurs is known as the ‘scissile bond’. In the Schechter and Berger nomenclature (14), residues on the left-hand side of the scissile bond (towards the N-terminus) are numbered P1, P2, P3, etc. and residues on the right-hand side (towards the C-terminus) are numbered P1′, P2′, P3′, etc. with cleavage occurring between P1 and P1′. The substrate-binding pocket in the peptidase that accommodates the P1 residue is known as the S1 pocket, and that accommodating the P1′ residue is the S1′ pocket.
Predicting where a peptidase will cleave in a native protein is difficult. Where cleavage does occur in a protein is due to a combination of the preference of the peptidase and the availability of bonds in the substrate. Although the preference of the peptidase can be quite simple, for example trypsin (MEROPS identifier S01.151) cleaves lysyl and arginyl bonds (15) and caspase-3 (C14.003) cleaves only aspartyl bonds (16), very often peptidase preference is cryptic. It is relatively easy to predict trypsin cleavages in a denatured protein, but few lysyl and arginyl bonds will be cleaved in a native protein. This has proved useful for researchers wishing to separate structural domains in a multidomain protein using limited proteolysis (17). It is not possible to predict where in a peptide cathepsin B (C01.060) will cleave, for example, despite its known preferences for a hydrophobic residue in the S2 pocket and arginine in S1 (18).
Even though for some peptidases the specificity has been clearly defined, in all probability only a few bonds will be susceptible to cleavage in a mature protein. A protein will have few bonds flexible enough to thread into a peptidase active site if the protein is in a native state, because of the stabilizing interactions within and between secondary structure elements within the substrate. It is widely assumed that the susceptible bonds will be within surface loops and interdomain connectors. However, once a bond is cleaved and the tertiary structure perturbed, further bonds may become susceptible.
Most studies of the action of a peptidase on a supposed physiological substrate are performed in vitro. It may be, however, that in vivo peptidase and substrate do not meet, either because of a physical boundary, such as being in different intracellular (or extracellular) compartments, because inhibitors inactivate the peptidase, the cleavage sites are inaccessible because the substrate is bound to another protein, or the environment is unsuitable and the peptidase is not active.
Despite the importance of protein cleavage, there has been no centralized repository for cleavage data collection and no attempt to curate these cleavages by mapping them to residue positions in protein primary sequence databases. Given that nearly all proteins are eventually degraded, and that any one protein can be degraded by several different peptidases often by cleavages at multiple peptide bonds, the potential total number of cleavages will always exceed the number of known proteins. Up until recently each cleavage had to be characterized biochemically, which meant N-terminal sequencing of the products, a time-consuming and labour-intensive task. Now that proteomic analyses are possible, where cell lysates or similar samples are subjected to cleavage by a peptidase, peptides isolated, composition determined by mass spectroscopy, and possible source protein(s) determined from the composition (19), the amount of data is set to rise exponentially. This makes it vitally important that the information be accurately stored and curated. Such a collection made readily available would provide a comprehensive training set for algorithms and software for the prediction of physiological substrates and cleavage positions.
The classification of peptidases into clans and families was first published in 1993 (20), and this was converted into an Internet resource, the MEROPS database (21), in 1996. The database was extended to include nomenclature and bibliographies, and has been developed over the years to be a one-stop shop for researchers with an interest in proteolysis. The collection of known cleavages in substrates which was started in 1998 (22) has now been added to the MEROPS database. For each peptidase there is a page listing known substrates, and, where enough substrates are known, the peptidase summary has displays to show peptidase specificity. For each protein substrate, the sequence is displayed showing where cleavage occurs and which peptidase performs that cleavage. In addition to the MEROPS collection, there is also a collection of physiologically relevant protein cleavages assembled by the CutDB database (23) and more specialist collections of substrates for individual peptidases or peptidase families, such as CASBAH for caspases (24).
Methods
Data collection and curation
The primary source of protein cleavage information is the published literature. Search profiles have been developed for use at PubMed (25) and Scopus (http://info.scopus.com/). These are updated regularly and currently include ∼500 names that are known to be used for peptidases. These retrieve a set of ∼250 potentially interesting abstracts each week. There is much redundancy, in that a single article may be retrieved by several search terms. Once a non-redundant list of articles has been obtained, the abstracts are reviewed to select the subset that is to be included in MEROPS. In a typical week, 50–60 references come through this filter. Keywords, including the MEROPS identifiers for the relevant peptidases, are manually attached to each and these determine which pages in MEROPS the reference will appear on. If from the abstract it is clear that the paper contains substrate-cleavage data these are entered immediately into the MEROPS collection. Periodically, the bibliography in MEROPS for a peptidase is reviewed to find substrate cleavages with a preference for peptidases without any substrates in the MEROPS collection.
If the substrate is a protein, it is mapped to a UniProt protein sequence database entry (26) initially by name and species. Each cleavage in the protein is mapped to a specific residue in the UniProt entry. Frequently the residue number reported in the paper refers to a position in the mature protein, and to map this to the UniProt sequence the length of any signal peptide and/or propeptide has to be added. The UniProt accession, the P1 residue number, the CRC64 checksum for the sequence and the MEROPS identifier for the peptidase are stored. In addition other information may be retained, including whether the cleavage is deemed by the authors of the source paper to be physiological or not, whether the substrate was in native conformation, the pH of the reaction, and the method used to identify the cleavage. The four residues either side of the scissile bond are also stored so that the cleavage position can be recalculated should the UniProt protein sequence change, and to provide the data for what amino acids are acceptable in the binding pockets S4–S4′ for each peptidase. A bespoke program (in Perl) was written to add each cleavage in a protein substrate to ensure consistency; the program connects to the locally installed version of UniProt so that each cleavage position can be confirmed as the data are entered. Some data were acquired from proteomics studies. Again a bespoke program was written to parse the data from the Excel spreadsheets available as Supplementary Data to the published papers. Some cleavages were acquired from the CutDB database, but these have been manually checked against the original reference and the UniProt sequence. Once again a bespoke program was written to collect the data, translate the provided substrate Protein Identifier to a Uniprot accession, check that a cleavage event was not already present in the MEROPS collection (and add the CutDB accession number if it were), and add new cleavage events to the MEROPS collection, reporting any inconsistency between the P4–P4′ residues and the sequence in the UniProt entry.
The data collected are non-redundant. If more than one paper reports the cleavage at the same position in the same protein by the same peptidase, then only data from the paper published first is retained. If several peptidases cleave the same protein in the same position, each is considered a different cleavage event and each is entered. There is no attempt to map cleavages to isoforms of a protein, unless different isoforms were used by the original researchers. Synthetic substrates that differ only in leader (for example benzyloxycarbonyl, succinyl or tosyl) or reporter groups (for example aminomethylcoumarin, naphthylamide or nitroanilide) are considered different cleavages even if all are performed by the same peptidase.
Specificity displays
The MEROPS website has two displays to show peptidase specificity. Both use the data from natural and synthetic substrates, but show only naturally occurring amino acids. The first display is a logo which uses the WebLogo software (27). Residues P4–P4′ from all the substrates for a peptidase are treated as an alignment. The observed frequency for each amino acid in each position is calculated as a bit score, the maximum possible being 4.32 bits. An amino acid is shown in the logo (in single-letter code) if the bit score exceeds 0.1. The logo is also shown as a text string, where if a single amino acid predominates at one position (i.e. the bit score exceeds 0.4) the letter is shown in uppercase, and if more than one amino acid predominates in any position a letter is shown in uppercase when the bit score exceeds 0.7 and in lower case if the bit score is between 0.1 and 0.7.
The second display is a frequency matrix, which is an 8 × 20 matrix with residues P4–P4′ along the x-axis and all amino acids along the y-axis. The amino acids are ordered so that those with similar properties are adjacent. The order is Gly, Pro, Ala, Val, Leu, Ile, Met, Phe, Tyr, Trp, Ser, Thr, Cys, Asn, Gln, Asp, Glu, Lys, Arg and His. Preference is calculated in terms of the percentage of substrates with each amino acid in each position, and a different shade of green is used for each tenth percentile interval. The number of times a residue occurs at each position is shown.
Sequence alignments
The UniProt accession for each substrate with a known cleavage site was used to search the UniRef50 database (clusters of sequences that have at least 50% sequence identity to the longest sequence) (28). If a UniRef50 entry was found, then all the UniProt accessions included in that entry were extracted and the sequences retrieved from the UniProt database in FastA format. Short fragments were excluded and the remaining sequences were then aligned with MUSCLE (29), using the default parameters and performing two iterations over the complete alignment to minimize gaps. Because each UniRef50 entry contains sequences sharing 50% or more sequence identity, the program is very quick, and the resulting alignment approximates to an alignment of orthologues. However, some UniRef50 entries will also contain closely related paralogues.
Sequence alignments were generated and highlighted to show not just conserved residues but also peptidase preference. For each cleavage the residues P4–P4′ were highlighted to indicate whether the residue in each sequence had been observed in any substrate at that position for the peptidase in question. Residues identical to the sequence where the cleavage is known are shown with a pink background. Replacements observed in other substrates are shown with an orange background. Where no substrate for this peptidase is known with this amino acid in this position the residue is shown with a black background. The term ‘atypical’ is used for an amino acid that has not been observed in a particular binding pocket in any known substrate for a peptidase.
Results and discussion
The MEROPS cleavage collection
The MEROPS cleavage collection contains 39 118 cleavages (as of 7 August 2009). The number of cleavages that can be mapped to entries in the UniProt database is 34 606, the remaining 4512 consisting mainly of synthetic substrates. The number of different entries in the UniProt database to which cleavages are mapped is 10513. The number of cleavages that are designated as ‘physiological’ is 13 307; whereas 20 187 cleavages are designated ‘non-physiological’ and 2677 cleavages are designated ‘synthetic’. The remaining 2947 are cleavages in peptides that can not be mapped to UniProt because: they are too short; they are significantly modified, such as the non-alpha peptide bond between ubiquitin and its target protein; they are derived from phage displays; they are theoretical cleavages or because it is unclear whether the cleavage is physiological or not. The data are derived from 6095 publications. The number of cleavages in common between the MEROPS and CutDB collections is 5876, of which 3424 were originally found in the literature by the CutDB researchers. The number of cleavages from the CutDB database that failed to make the MEROPS collection, excluding 892 isoforms and 35 duplicates, was 560 (9.5%), mostly due to being mapped to the wrong residue or sequence. The CutDB curators have been informed of these discrepancies. Because the CutDB database includes only cleavages thought to be of physiological relevance and those that can be included in their proteolytic pathways, it has fewer cleavages than in the MEROPS collection. It does not include cleavages in synthetic substrates and those peptides used solely to map peptidase specificities, or general purpose processing enzymes such as signal peptidases and methionyl aminopeptidases.
There are 2415 different peptidases recognized in the MEROPS database (excluding hypothetical peptidases from model organisms). Substrate cleavages have been collected for 1086 peptidases (45%); for the remainder any cleavage positions in substrates are either unknown or have not yet been found in the literature. Only 312 peptidases have had ten or more cleavages collected, and it is only these for which there is enough data for further analysis. The total number of cleavages for these 312 peptidases is 33 047. The peptidases with most cleavage data collected are (the MEROPS identifier and number of cleavages are given in parenthesis after the name): trypsin 1 (S01.151; 13 558), matrix metallopeptidase-2 (M10.003; 2227), eukaryotic signal peptidase complex (XS26-001; 1801), glutamyl peptidase I (S01.269; 1213), HIV-1 retropepsin (A02.001; 1059), methionyl aminopeptidase 1 (M24.001; 564), cathepsin G (S01.133; 448), chymotrypsin A (cattle-type) (S01.001; 445), caspase-3 (C14.003; 414), elastase-2 (S01.131; 400), signalase (animal) 21 kDa component (S26.010; 363) and granzyme B (Homo sapiens-type) (S01.010; 358).
Peptidase specificity
The residues from P4–P4′ were collected for each substrate cleavage. Figure 1 shows the number of peptidases showing some selectivity for one or two residues in each binding pocket from S4 to S4′. Clearly, many peptidases have extended substrate-binding sites with preferences beyond S1, with 52 showing a preference in the S4 pocket. There are a few peptidases that have a preference at S5 (30), but a preference so far from the scissile bond is rare. It is conceivable that mitochondrial intermediate peptidase (M03.006), which removes a transit octapeptide from the N-terminus of proteins synthesized in the cytoplasm but destined for the mitochondrial matrix, may have a preference as far away from the cleavage site as S8 (31). Preference on the prime side of the scissile bond is thought to rarely extend beyond the S1′ pocket, but Figure 1 shows that 32 different peptidases have a preference in S4′.
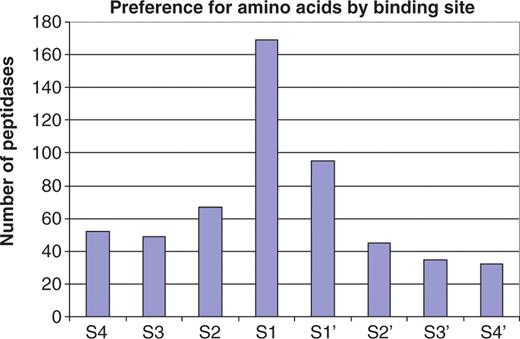
Preference for amino acids in substrate binding sites. The bar chart shows the number of peptidases showing a preference for one or two amino acids for each substrate binding site S4–S4′. Of the 312 peptidase with 10 or more known substrate cleavages, 202 show a preference and are included in the figure. A count is made whenever an amino acid occurs in one binding pocket in 40% or more of the substrates. There are 15 peptidases that have a preference for two amino acids in a binding pocket: walleye dermal sarcoma virus retropepsin (A02.063, Asn or Gln in S2), sapovirus 3C-like peptidase (C24.003, Glu or Gln in S1), SARS coronavirus picornain 3C-like peptidase (C30.005, Gly or Gln in S1), peptidyl-peptidase Acer (M02.002, Gly or Pro in S1), vimelysin (M04.010, Phe or Leu in S1), carboxypeptidase M (M14.006, Arg or Lys in S1′), carboxypeptidase U (M14.009, Arg or Lys in S1′), dactylysin (M9G.026, Leu or Phe in S1′), chymase (S01.140, Phe or Tyr in S1), tryptase alpha (S01.143, Lys or Arg in S1), trypsin 1 (S01.151, Lys or Arg in S1), plasmin (S01.233, Lys or Arg in S1), flavivirin (S07.001, Lys or Arg in S2), dipeptidyl aminopeptidase A (S09.005, Ala or Pro in S1) and kumamolisin (S53, 004, Glu or Gly in S3). Many peptidases show a preference in more than one binding pocket. There are 13 peptidases with a preference for all eight binding pockets, another 13 with a preference in seven, five peptidases in six, three in five, eight in four, 24 in three, 47 in two and 89 in only one.
Exopeptidases cleave near protein termini, and because the binding pockets do not exist are unable to accept any amino acids in some positions. Dipeptidases can only accept residues in the S1 and S1′ pockets; aminopeptidases are unable to accept any residue in S4–S2, carboxypeptidases in S2′–S4′, dipeptidyl-peptidases in S4 and S3, tripeptidyl-peptidases in S4 and peptidyl-dipeptidases in S3′ and S4′. Some omega peptidases (peptidases which do not cleave normal peptide bonds but release substituted amino acids such as pyroglutamate or cleave isopeptide bonds, such as many deubiquitinating enzymes) may also be unable to accept any residue in certain positions, or it is not possible to interpret cleavages in terms of P4–P4′, for example for isopeptidases. There are 36 peptidases with 10 or more cleavages that cannot accept any residue in S4, 35 for S3, 26 for S2, 15 for S2′, 22 for S3′ and 25 for S4′.
Table 1 shows the number of peptidases showing some selectivity in each binding pocket from S4 to S4′ for amino acid properties (where ‘acidic’ is Asp or Glu; ‘basic’ is Arg, His or Lys; ‘aliphatic’ is Ile, Leu or Val; ‘aromatic’ is Phe, Trp or Tyr; and ‘small’ is Ala, Cys, Gly or Ser). Properties are taken from Livingstone and Barton (32). Only the categories with the fewest amino acids, and those that do not overlap (with the exception of His, which can also be considered aromatic) have been used. If categories such as ‘hydrophobic’ and ‘polar’ are used then nearly every binding pocket is highlighted because each category contains more than half of the amino acids. Most preference is directed towards the S1 (201 different peptidases) and S1′ (160 different peptidases) pockets. The commonest preferences are for a basic amino acid in the P1 position, small amino acids in P1 and P1′, and an aliphatic amino acid in P1′. No aromatic amino acids were observed in P4′ in any of the substrates of these 312 peptidases. For each amino acid category preference was most pronounced in the S1 pocket with the exception of aliphatic amino acids, where most peptidases have a preference in the S1′ pocket. Preference for acidic amino acids is very rare except in the S1 pocket, and similarly aromatic amino acids are rarely preferred except in S1 and S1′.
| Amino acid type . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Acidic | 5 | 7 | 5 | 24 | 5 | 4 | 2 | 5 |
| Basic | 11 | 8 | 13 | 67 | 11 | 9 | 5 | 2 |
| Aliphatic | 22 | 24 | 32 | 18 | 56 | 36 | 23 | 7 |
| Aromatic | 2 | 2 | 8 | 34 | 23 | 7 | 1 | 0 |
| Small | 35 | 34 | 31 | 58 | 65 | 22 | 26 | 20 |
| Total | 75 | 75 | 89 | 201 | 160 | 78 | 57 | 34 |
| Amino acid type . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Acidic | 5 | 7 | 5 | 24 | 5 | 4 | 2 | 5 |
| Basic | 11 | 8 | 13 | 67 | 11 | 9 | 5 | 2 |
| Aliphatic | 22 | 24 | 32 | 18 | 56 | 36 | 23 | 7 |
| Aromatic | 2 | 2 | 8 | 34 | 23 | 7 | 1 | 0 |
| Small | 35 | 34 | 31 | 58 | 65 | 22 | 26 | 20 |
| Total | 75 | 75 | 89 | 201 | 160 | 78 | 57 | 34 |
The number of peptidases with a preference for a particular amino acid type for each binding pocket S4–S4′ is shown, where 40% or more of substrates have an amino acid of that type at that position. Only those 312 peptidases with at least 10 known cleavages are included. There are 276 peptidases that show a preference, of which 18 show a preference at all eight sites, 16 for seven sites, 12 for six sites, 17 for five sites, 33 for four sites, 46 for three sites, 64 for two sites and 70 for one site.
| Amino acid type . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Acidic | 5 | 7 | 5 | 24 | 5 | 4 | 2 | 5 |
| Basic | 11 | 8 | 13 | 67 | 11 | 9 | 5 | 2 |
| Aliphatic | 22 | 24 | 32 | 18 | 56 | 36 | 23 | 7 |
| Aromatic | 2 | 2 | 8 | 34 | 23 | 7 | 1 | 0 |
| Small | 35 | 34 | 31 | 58 | 65 | 22 | 26 | 20 |
| Total | 75 | 75 | 89 | 201 | 160 | 78 | 57 | 34 |
| Amino acid type . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Acidic | 5 | 7 | 5 | 24 | 5 | 4 | 2 | 5 |
| Basic | 11 | 8 | 13 | 67 | 11 | 9 | 5 | 2 |
| Aliphatic | 22 | 24 | 32 | 18 | 56 | 36 | 23 | 7 |
| Aromatic | 2 | 2 | 8 | 34 | 23 | 7 | 1 | 0 |
| Small | 35 | 34 | 31 | 58 | 65 | 22 | 26 | 20 |
| Total | 75 | 75 | 89 | 201 | 160 | 78 | 57 | 34 |
The number of peptidases with a preference for a particular amino acid type for each binding pocket S4–S4′ is shown, where 40% or more of substrates have an amino acid of that type at that position. Only those 312 peptidases with at least 10 known cleavages are included. There are 276 peptidases that show a preference, of which 18 show a preference at all eight sites, 16 for seven sites, 12 for six sites, 17 for five sites, 33 for four sites, 46 for three sites, 64 for two sites and 70 for one site.
The preference for individual amino acids is shown in Table 2. It is clear from the table that cysteine is an unwelcome amino acid near a cleavage site. Only the peroxisomal transit peptide peptidase shows a preference for cysteine binding to the S2 and S1 pockets. Tryptophan is also rare around cleavage sites, with only tryptophanyl aminopeptidase (M9A.008, a preference in the S1 pocket) and mast cell peptidase 4 (Rattus) (S01.005, in the S2 pocket) showing a preference; however, this may have more to do with the fact that tryptophan is the rarest of the amino acids. Asparagine is also very rare in the proximity of a cleavage site, one of the few examples being the specialist peptidase legumain (C13.006) which only cleaves asparaginyl bonds (33). Histidine is also a rare preference, with only three peptidases showing any preference for it, namely chymosin (A01.006; S4), carnosine dipeptidase I (M20.006; S1′) and Xaa-methyl-His dipeptidase (M20.013; S1′). Methionine is also not preferred by most peptidases, exceptions being methionyl aminopeptidases (M24.001, M24.002), where the preference is as expected for methionine binding in the S1 pocket, some members of the peptidase Clp family (S14) and the unsequenced Met-Xaa dipeptidase (M9B.004). The gpr peptidase (A25.001) shows a preference for Met binding to S4. The commonest preference is for arginine binding to the S1 pocket, which occurs in over fifty peptidases. However, arginine is relatively rare outside the P1 position. There are peptidases that show a preference for Gly, Pro and Val for every binding pocket in the range S4–S4′. Peptidases showing unique preferences are listed in Table 3.
| Amino acid . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Ala | 6 | 8 | 5 | 10 | 8 | 5 | 1 | |
| Cys | 1 | 1 | ||||||
| Asp | 3 | 16 | 2 | 3 | ||||
| Glu | 1 | 7 | 5 | 8 | 1 | 1 | 2 | |
| Phe | 2 | 1 | 5 | 12 | 10 | 2 | ||
| Gly | 3 | 1 | 11 | 17 | 12 | 2 | 6 | 5 |
| His | 1 | 2 | ||||||
| Ile | 2 | 1 | 8 | 1 | ||||
| Lys | 2 | 4 | 8 | 6 | 6 | 2 | 4 | |
| Leu | 11 | 4 | 9 | 12 | 24 | 4 | 7 | |
| Met | 1 | 6 | ||||||
| Asn | 9 | 1 | ||||||
| Pro | 2 | 8 | 5 | 9 | 9 | 4 | 1 | 4 |
| Gln | 9 | 1 | 5 | 1 | 10 | |||
| Arg | 8 | 1 | 2 | 52 | 5 | 3 | 1 | |
| Ser | 8 | 1 | 8 | 3 | 2 | 1 | ||
| Thr | 3 | 1 | 1 | 1 | ||||
| Val | 1 | 6 | 1 | 2 | 5 | 6 | 11 | 5 |
| Trp | 1 | 1 | ||||||
| Tyr | 11 | 1 | 5 |
| Amino acid . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Ala | 6 | 8 | 5 | 10 | 8 | 5 | 1 | |
| Cys | 1 | 1 | ||||||
| Asp | 3 | 16 | 2 | 3 | ||||
| Glu | 1 | 7 | 5 | 8 | 1 | 1 | 2 | |
| Phe | 2 | 1 | 5 | 12 | 10 | 2 | ||
| Gly | 3 | 1 | 11 | 17 | 12 | 2 | 6 | 5 |
| His | 1 | 2 | ||||||
| Ile | 2 | 1 | 8 | 1 | ||||
| Lys | 2 | 4 | 8 | 6 | 6 | 2 | 4 | |
| Leu | 11 | 4 | 9 | 12 | 24 | 4 | 7 | |
| Met | 1 | 6 | ||||||
| Asn | 9 | 1 | ||||||
| Pro | 2 | 8 | 5 | 9 | 9 | 4 | 1 | 4 |
| Gln | 9 | 1 | 5 | 1 | 10 | |||
| Arg | 8 | 1 | 2 | 52 | 5 | 3 | 1 | |
| Ser | 8 | 1 | 8 | 3 | 2 | 1 | ||
| Thr | 3 | 1 | 1 | 1 | ||||
| Val | 1 | 6 | 1 | 2 | 5 | 6 | 11 | 5 |
| Trp | 1 | 1 | ||||||
| Tyr | 11 | 1 | 5 |
The number of peptidases showing a preference for an amino acid in a binding site is shown. Only those 312 peptidases with 10 or more known substrate cleavages are included. An amino acid must occur at that position in 40% or more of substrates. Therefore, it is possible for two amino acids to be preferred in any one binding pocket, as is the case for trypsin 1 where there is a preference for either Lys (59% of substrates) or Arg (41%) in S1. There are 202 peptidases that show a preference, of which 13 show a preference at all eight sites, 13 for seven sites, five for six sites, three for five sites, eight for four sites, 23 for three sites, 49 for two sites and 88 for one site.
| Amino acid . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Ala | 6 | 8 | 5 | 10 | 8 | 5 | 1 | |
| Cys | 1 | 1 | ||||||
| Asp | 3 | 16 | 2 | 3 | ||||
| Glu | 1 | 7 | 5 | 8 | 1 | 1 | 2 | |
| Phe | 2 | 1 | 5 | 12 | 10 | 2 | ||
| Gly | 3 | 1 | 11 | 17 | 12 | 2 | 6 | 5 |
| His | 1 | 2 | ||||||
| Ile | 2 | 1 | 8 | 1 | ||||
| Lys | 2 | 4 | 8 | 6 | 6 | 2 | 4 | |
| Leu | 11 | 4 | 9 | 12 | 24 | 4 | 7 | |
| Met | 1 | 6 | ||||||
| Asn | 9 | 1 | ||||||
| Pro | 2 | 8 | 5 | 9 | 9 | 4 | 1 | 4 |
| Gln | 9 | 1 | 5 | 1 | 10 | |||
| Arg | 8 | 1 | 2 | 52 | 5 | 3 | 1 | |
| Ser | 8 | 1 | 8 | 3 | 2 | 1 | ||
| Thr | 3 | 1 | 1 | 1 | ||||
| Val | 1 | 6 | 1 | 2 | 5 | 6 | 11 | 5 |
| Trp | 1 | 1 | ||||||
| Tyr | 11 | 1 | 5 |
| Amino acid . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|
| Ala | 6 | 8 | 5 | 10 | 8 | 5 | 1 | |
| Cys | 1 | 1 | ||||||
| Asp | 3 | 16 | 2 | 3 | ||||
| Glu | 1 | 7 | 5 | 8 | 1 | 1 | 2 | |
| Phe | 2 | 1 | 5 | 12 | 10 | 2 | ||
| Gly | 3 | 1 | 11 | 17 | 12 | 2 | 6 | 5 |
| His | 1 | 2 | ||||||
| Ile | 2 | 1 | 8 | 1 | ||||
| Lys | 2 | 4 | 8 | 6 | 6 | 2 | 4 | |
| Leu | 11 | 4 | 9 | 12 | 24 | 4 | 7 | |
| Met | 1 | 6 | ||||||
| Asn | 9 | 1 | ||||||
| Pro | 2 | 8 | 5 | 9 | 9 | 4 | 1 | 4 |
| Gln | 9 | 1 | 5 | 1 | 10 | |||
| Arg | 8 | 1 | 2 | 52 | 5 | 3 | 1 | |
| Ser | 8 | 1 | 8 | 3 | 2 | 1 | ||
| Thr | 3 | 1 | 1 | 1 | ||||
| Val | 1 | 6 | 1 | 2 | 5 | 6 | 11 | 5 |
| Trp | 1 | 1 | ||||||
| Tyr | 11 | 1 | 5 |
The number of peptidases showing a preference for an amino acid in a binding site is shown. Only those 312 peptidases with 10 or more known substrate cleavages are included. An amino acid must occur at that position in 40% or more of substrates. Therefore, it is possible for two amino acids to be preferred in any one binding pocket, as is the case for trypsin 1 where there is a preference for either Lys (59% of substrates) or Arg (41%) in S1. There are 202 peptidases that show a preference, of which 13 show a preference at all eight sites, 13 for seven sites, five for six sites, three for five sites, eight for four sites, 23 for three sites, 49 for two sites and 88 for one site.
| Peptidase name . | MEROPS ID . | Total substrate cleavages . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|---|---|
| Chymosin | A01.006 | 15 | His | Ser | Ile | |||||
| Feline immunodefiency virus retropepsin | A02.007 | 28 | Val | |||||||
| Walleye dermal sarcoma virus retropepsin | A02.063 | 27 | Gln | |||||||
| PibD peptidase | A24.017 | 10 | Thr | |||||||
| gpr peptidase | A25.001 | 32 | Met | Ile | Glu | |||||
| Cruzipain | C01.075 | 49 | Arg | |||||||
| Coxsackievirus-type picornain 3C | C03.011 | 10 | Pro | |||||||
| Ubiquitinyl hydrolase-L3 | C12.003 | 14 | Arg | |||||||
| Legumain | C13.004 | 30 | Asn | |||||||
| Sapovirus 3C-like peptidase | C24.003 | 10 | Thr | |||||||
| Separase (yeast-type) | C50.001 | 12 | Glu | |||||||
| Peptidyl-dipeptidase Acer | M02.002 | 10 | Phe | |||||||
| Bacterial collagenase H | M09.003 | 18 | Ala | |||||||
| PrtA peptidase (Photorhabdus-type) | M10.063 | 23 | Glu | |||||||
| ADAM8 peptidase | M12.208 | 22 | Gln | |||||||
| Tryptophanyl aminopeptidase (Trichosporon cutaneum) | M9A.008 | 15 | Trp | |||||||
| Carboxypeptidase G3 | M9E.007 | 12 | Glu | |||||||
| Mast cell peptidase 4 (Rattus) | S01.005 | 33 | Trp | |||||||
| Kumamolisin | S53.004 | 10 | Val | Gly | Tyr | |||||
| Peroxisomal transit peptide peptidase | U9G.062 | 14 | Cys | Cys |
| Peptidase name . | MEROPS ID . | Total substrate cleavages . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|---|---|
| Chymosin | A01.006 | 15 | His | Ser | Ile | |||||
| Feline immunodefiency virus retropepsin | A02.007 | 28 | Val | |||||||
| Walleye dermal sarcoma virus retropepsin | A02.063 | 27 | Gln | |||||||
| PibD peptidase | A24.017 | 10 | Thr | |||||||
| gpr peptidase | A25.001 | 32 | Met | Ile | Glu | |||||
| Cruzipain | C01.075 | 49 | Arg | |||||||
| Coxsackievirus-type picornain 3C | C03.011 | 10 | Pro | |||||||
| Ubiquitinyl hydrolase-L3 | C12.003 | 14 | Arg | |||||||
| Legumain | C13.004 | 30 | Asn | |||||||
| Sapovirus 3C-like peptidase | C24.003 | 10 | Thr | |||||||
| Separase (yeast-type) | C50.001 | 12 | Glu | |||||||
| Peptidyl-dipeptidase Acer | M02.002 | 10 | Phe | |||||||
| Bacterial collagenase H | M09.003 | 18 | Ala | |||||||
| PrtA peptidase (Photorhabdus-type) | M10.063 | 23 | Glu | |||||||
| ADAM8 peptidase | M12.208 | 22 | Gln | |||||||
| Tryptophanyl aminopeptidase (Trichosporon cutaneum) | M9A.008 | 15 | Trp | |||||||
| Carboxypeptidase G3 | M9E.007 | 12 | Glu | |||||||
| Mast cell peptidase 4 (Rattus) | S01.005 | 33 | Trp | |||||||
| Kumamolisin | S53.004 | 10 | Val | Gly | Tyr | |||||
| Peroxisomal transit peptide peptidase | U9G.062 | 14 | Cys | Cys |
| Peptidase name . | MEROPS ID . | Total substrate cleavages . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|---|---|
| Chymosin | A01.006 | 15 | His | Ser | Ile | |||||
| Feline immunodefiency virus retropepsin | A02.007 | 28 | Val | |||||||
| Walleye dermal sarcoma virus retropepsin | A02.063 | 27 | Gln | |||||||
| PibD peptidase | A24.017 | 10 | Thr | |||||||
| gpr peptidase | A25.001 | 32 | Met | Ile | Glu | |||||
| Cruzipain | C01.075 | 49 | Arg | |||||||
| Coxsackievirus-type picornain 3C | C03.011 | 10 | Pro | |||||||
| Ubiquitinyl hydrolase-L3 | C12.003 | 14 | Arg | |||||||
| Legumain | C13.004 | 30 | Asn | |||||||
| Sapovirus 3C-like peptidase | C24.003 | 10 | Thr | |||||||
| Separase (yeast-type) | C50.001 | 12 | Glu | |||||||
| Peptidyl-dipeptidase Acer | M02.002 | 10 | Phe | |||||||
| Bacterial collagenase H | M09.003 | 18 | Ala | |||||||
| PrtA peptidase (Photorhabdus-type) | M10.063 | 23 | Glu | |||||||
| ADAM8 peptidase | M12.208 | 22 | Gln | |||||||
| Tryptophanyl aminopeptidase (Trichosporon cutaneum) | M9A.008 | 15 | Trp | |||||||
| Carboxypeptidase G3 | M9E.007 | 12 | Glu | |||||||
| Mast cell peptidase 4 (Rattus) | S01.005 | 33 | Trp | |||||||
| Kumamolisin | S53.004 | 10 | Val | Gly | Tyr | |||||
| Peroxisomal transit peptide peptidase | U9G.062 | 14 | Cys | Cys |
| Peptidase name . | MEROPS ID . | Total substrate cleavages . | S4 . | S3 . | S2 . | S1 . | S1′ . | S2′ . | S3′ . | S4′ . |
|---|---|---|---|---|---|---|---|---|---|---|
| Chymosin | A01.006 | 15 | His | Ser | Ile | |||||
| Feline immunodefiency virus retropepsin | A02.007 | 28 | Val | |||||||
| Walleye dermal sarcoma virus retropepsin | A02.063 | 27 | Gln | |||||||
| PibD peptidase | A24.017 | 10 | Thr | |||||||
| gpr peptidase | A25.001 | 32 | Met | Ile | Glu | |||||
| Cruzipain | C01.075 | 49 | Arg | |||||||
| Coxsackievirus-type picornain 3C | C03.011 | 10 | Pro | |||||||
| Ubiquitinyl hydrolase-L3 | C12.003 | 14 | Arg | |||||||
| Legumain | C13.004 | 30 | Asn | |||||||
| Sapovirus 3C-like peptidase | C24.003 | 10 | Thr | |||||||
| Separase (yeast-type) | C50.001 | 12 | Glu | |||||||
| Peptidyl-dipeptidase Acer | M02.002 | 10 | Phe | |||||||
| Bacterial collagenase H | M09.003 | 18 | Ala | |||||||
| PrtA peptidase (Photorhabdus-type) | M10.063 | 23 | Glu | |||||||
| ADAM8 peptidase | M12.208 | 22 | Gln | |||||||
| Tryptophanyl aminopeptidase (Trichosporon cutaneum) | M9A.008 | 15 | Trp | |||||||
| Carboxypeptidase G3 | M9E.007 | 12 | Glu | |||||||
| Mast cell peptidase 4 (Rattus) | S01.005 | 33 | Trp | |||||||
| Kumamolisin | S53.004 | 10 | Val | Gly | Tyr | |||||
| Peroxisomal transit peptide peptidase | U9G.062 | 14 | Cys | Cys |
Despite there being a large number of substrates collected, the specificity of some peptidases can not be explained in terms of S4–S4′ preferences. These peptidases include (MEROPS identifier and number of substrate cleavages in brackets): cathepsin D (A01.009; 145), cathepsin E (A01.010; 64), nemepsin-2 (A01.068; 127), papain (C01.001; 40), cathepsin X (C01.013; 24), cathepsin L (C01.032; 85), cathepsin B (C01.060; 82), aspergilloglutamic peptidase (G01.002; 37), mirabilysin (M10.057; 32), neprilysin (M13.001; 83), endothelin-converting enzyme 1 (M13.002; 27), MEP peptidase (M13.011; 43), pitrilysin (M16.001; 23), insulysin (M16.002; 31), eupitrilysin (M16.009; 54), aminopeptidase Ap1 (M28.002; 66), plasma glutamate carboxypeptidase (M28.014; 33), penicillolysin (M35.001; 20), deuterolysin (M35.002; 22), FtsH peptidase (M41.001; 24), dipeptidyl-peptidase III (M49.001; 24), glycyl aminopeptidase (M61.001; 26), chymotrypsin C (S01.157; 20), kallikrein 1 (S01.160; 25), subtilisin Carlsberg (S08.001; 33), high alkaline protease (Alkaliphilus transvaalensis) (S08.028; 28), peptidase K (S08.054; 43) and signalase (animal) 21 kDa component (S26.010; 363).
Displays on the MEROPS website
Specificity logos and frequency matrices present the user with a visual representation of peptidase specificity. An example specificity logo is shown in Figure 2. From the logo and the cleavage pattern string it is clear that caspase-3 has an absolute requirement for Asp in the S1 pocket (position 4, only one cleavage after Glu is known) and a preference for Asp in S4. There are minor preferences for Glu in S3 and Gly or Ser in S1′.
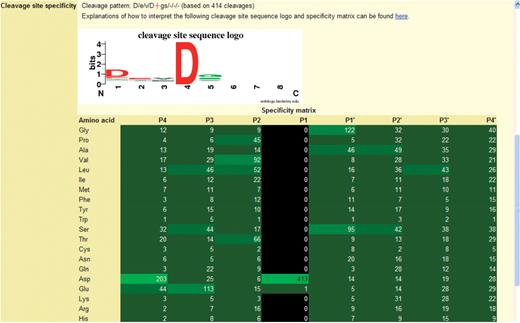
The specificity logo and frequency matrix showing the substrate specificity of caspase-3. The figure is taken from a page in the MEROPS database. The logo is shown at the top with the frequency matrix below. The cleavage pattern is a textual representation of the logo, where the scissile bond is shown as a red cross, and the binding pockets separated by forward slashes. The preferred residue is shown in uppercase if the preference is strong. The number of cleavages on which these data are based is given in parentheses. For the logo, the binding pockets S4–S4′ are shown along the x-axis, where 1 is S4, 2 is S3, etc. The bit score is shown on the y-axis. The height of the letter is proportional to the bit score. The letters are coloured to indicate amino acid properties: blue for basic, red for acidic, black for hydrophobic and green for any other. In the frequency matrix below the logo, each cell shows the number of substrates with an amino acid occupying one of the positions P4–P4′. Cells in the matrix are highlighted in shades of green where the greater the preference, i.e. the more often an amino acid occurs at that position, the brighter the shade. Cells are highlighted in black if the amino acid is unknown at that position for any substrate.
While the logo indicates which amino acids are acceptable in each position, it does not indicate which amino acids are unobserved. These are shown in the frequency matrix, and an example is also shown in Figure 2 for caspase-3. In this example Asp occurs in the P1 position in all 413 substrates, Asp occurs in P4 in almost half the substrates, while Glu occurs in P3 in 27% of substrates. Note that in this frequency matrix every amino acid occurs in positions P4–P2 and P1′–P4′, but tryptophan is observed only once in P4, P2, P1′ and P4′. This gives an indication of the minimum number of substrate cleavages that has to be collected for a peptidase before definite conclusions about specificity in all binding pockets can be drawn.
A substrate alignment is shown in Figure 3. The density of residues highlighted in black is high, implying that this cleavage position is very poorly conserved and thus may not be physiologically relevant.
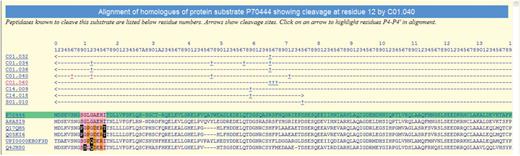
Alignment of the protein sequences of orthologues of the mouse BID protein showing known peptidase cleavages. The alignment is highlighted to show conservation of residues around the cleavage of BID by cathepsin H (C01.040) at residue 12. The sequence where the cleavage is known is highlighted in green and residues are numbered according to this sequence (inserts are indicated by letters). The rows beneath the residue numbers show the MEROPS identifier of each peptidase known to cleave this substrate. Arrows indicate the residue range of the fragment used in the experiment, and cleavage positions are indicated by the ‘+’ symbol. Clicking on a MEROPS identifier takes the user to the relevant summary page. Clicking on a ‘+’ symbol causes the alignment to be redrawn with residues P4–P4′ highlighted for that particular cleavage. Residues either side of the cleavage site are highlighted in pink if conserved with the equivalent residue in the sequence where the cleavage is known. A residue is highlighted in orange if it is not conserved but is known to occur in the same binding pocket in another cathepsin H substrate. A residue is shown as white on black if it is not conserved and is not known to occur in the same peptidase substrate binding site in any other substrate.
Substrate cleavages that are not evolutionarily conserved
Protein sequence alignments were constructed for every substrate where the cleavage had been assumed in the literature to be of physiological significance. The total number of alignments generated was 3141. A selection of cleavage sites which were not conserved in all homologues included in the same UniRef50 database entry are listed in Table 4. Only those cleavages by peptidases with at least 20 known substrates are included.
Assumed physiological cleavages that are not conserved in terms of peptidase substrate binding
| Substrate . | UniProt accession . | P1 . | Peptidase [MEROPS ID] (total substrates) . | Replacements . | Possible cause . | Ref. . |
|---|---|---|---|---|---|---|
| Serine protease HTRA2, mitochondrial | O43464 | 211 | HtrA2 peptidase [S01.278] (56) | VRLLSGDT (5) | (37) | |
| –-P–– (4) | g | |||||
| Cytochrome C | P00022 | 1 | mitochondrial methionyl aminopeptidase [M24.028] (131) | MGDVE (35) | (38) | |
| -C–- (1) | g | |||||
| Coagulation factor XIII A chain | P00488 | 38 | thrombin [S01.217] (169) | VVPRGVNL (22) | (39) | |
| –-L–– (1) | g | |||||
| Insulin-1 | P01325 | 87 | proprotein convertase 2 [S08.073] (59) | RQKRGIVD (36) | (40) | |
| –WH-W-W (1) | a | |||||
| –A-X–R (1) | d | |||||
| Collagen alpha-2(I) chain | P02465 | 870 | cathepsin D [A01.009] (145) | APGFLGLP (15) | (41) | |
| –-I–– (21) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−1 [M10.001] (70) | GPQGLLGA (28) | (42) | |
| -T––– (8) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−8 [M10.002] (87) | GPQGLLGA (28) | (42) | |
| -T–P–- (8) | h | |||||
| Platelet-derived growth factor subunit A | P04085 | 86 | Furin [S08.071] (116) | RRKRSIEE (38) | (43) | |
| G-LT–– (1) | b | |||||
| L–X–– (1) | d | |||||
| Collagen alpha-2(IV) chain | P08572 | 1077 | kallikrein-related peptidase 14 [S01.029] (49) | APGRAGLY (6) | (44) | |
| –-S–– (7) | a | |||||
| –-L–– (5) | a | |||||
| –-A–– (2) | a | |||||
| –-I–– (1) | a, g | |||||
| –-V–– (3) | ||||||
| Collagen alpha-2(IV) chain | P08572 | 1109 | kallikrein-related peptidase 14 [S01.029] (49) | KGERGTTG (12) | (44) | |
| –QP-E– (7) | a | |||||
| ––-E– (2) | a | |||||
| –-L–– (2) | a | |||||
| –-V–– (1) | a | |||||
| Insulin-like growth factor-binding protein 1 | P08833 | 165 | Matriptase [S01.302] (26) | KALHVTNI (2) | (45) | |
| –D-N–- (1) | b | |||||
| -SXXXDD- (1) | d | |||||
| –V––- (2) | h | |||||
| –-E–D- (3) | h | |||||
| Acyl-CoA thioesterase I | P0ADA1 | 26 | Signal peptidase I [S26.001] (294) | RAAAADTL (19) | (46) | |
| X–––- (1) | d | |||||
| Protein ygiW | P0ADU5 | 20 | Signal peptidase I [S26.001] (294) | PVMAAEQG (10) | (47) | |
| ––-X– (1) | d | |||||
| Chymotrypsin inhibitor 3 | P10822 | 24 | Signalase (animal) 21 kDa component [S26.010) (363) | SSTADDDL (4) | (48) | |
| X–––- (7) | d | |||||
| –-M–– (1) | h | |||||
| Plastocyanin minor isoform, chloroplastic | P11490 | 72 | Thylakoidal processing peptidase [S26.008] (52) | NAMAMEVL (20) | (49) | |
| ––Q–- (2) | h | |||||
| –-D–– (1) | g | |||||
| 50S ribosomal protein L7Ae | P12743 | 1 | Methionyl aminopeptidase 2 [M24.002] (130) | MPVYV (2) | (50) | |
| KKMA–– (1) | d | |||||
| SKDK–– (8) | d | |||||
| MAR–- (1) | d | |||||
| Beta-crystallin B3 | P19141 | 4 | Calpain-1 [C02.001] (101) | MAEQHSTP (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| Beta-crystallin B3 | P19141 | 10 | Calpain-1 [C02.001] (101) | TPEQAAAG (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | P19174 | 770 | Caspase-7 [C14.004] (112) | AEPDYGAL (20) | (52) | |
| T–––- (1) | g | |||||
| Mimecan | P20774 | 219 | ADAMTS4 peptidase [M12.221] (57) | TFLYLDHN (26) | (53) | |
| -H––– (3) | h | |||||
| Mimecan | P20774 | 234 | ADAMTS4 peptidase [M12.221] (57) | NLPESLRV (23) | (53) | |
| X–––- (6) | d | |||||
| Trypsin inhibitor 2 | P26780 | 30 | Signalase (animal) 21 kDa component [S26.010] (363) | IKAQDSEC (7) | (54) | |
| –-H–– (2) | a | |||||
| 60S ribosomal protein L10 | P27635 | 180 | Granzyme B (Homo sapiens)-type) [S01.010] (348) | NADEFEDM (36) | (55) | |
| R–––- (2) | ||||||
| Chitinase 2 | P29027 | 22 | Signalase (animal) 21 kDa component [S26.010] (363) | GVQAAWSS (2) | (56) | |
| XX––– (1) | a, d | |||||
| Alpha-synuclein | P37840 | 122 | Calpain-1 | DPDNEAYE (34) | (57) | |
| [C02.001] (101) | –-D–– (1) | g | ||||
| –X––- (2) | b, d | |||||
| Cathepsin E | P43159 | 53 | Cathepsin E [A01.010] (64) | KVDMVQYT (14) | (35) | |
| -Y––– (11) | a | |||||
| -F––– (2) | h | |||||
| -H––– (3) | a | |||||
| –-G–– (2) | a | |||||
| –-T–– (2) | h | |||||
| ––H–- (5) | a | |||||
| –––H- (1) | g | |||||
| 40S ribosomal protein S25 | P62852 | 51 | Granzyme B, | LFDKATYD (9) | (55) | |
| rodent-type [S01.136] (231) | –XXXXX- (2) | b, d | ||||
| Hemoglobin subunit alpha | P69905 | 37 | Cathepsin D [A01.009] (145) | FLSFPTTK (42) | (34) | |
| ––-W– (1) | h | |||||
| Hemoglobin subunit alpha | P69905 | 109 | Cathepsin D [A01.009] (145) | LLVTLAAH (36) | (34) | |
| –––C- (6) | h | |||||
| Hemoglobin subunit alpha | P69905 | 110 | Cathepsin D [A01.009] (145) | LVTLAAHL (36) | (34) | |
| ––-C– (6) | h | |||||
| ABC transporter periplasmic-binding protein yphF | P77269 | 26 | Signal peptidase I [S26.001] (294) | FARAAEKE (22) | (47) | |
| –-T–– (1) | g | |||||
| Tyrosine-protein phosphatase non-receptor type 18 | Q61152 | 424 | Caspase-1 [C14.001] (60) | EVTDGAQT (4) | (58) | |
| –-G–– (4) | h | |||||
| ––R–- (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 810 | ADAMTS5 peptidase [M12.225] (38) | ALVTATLG (14) | (53) | |
| -H–-N– (1) | h | |||||
| ––-N– (2) | h | |||||
| –––M- (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 811 | ADAMTS5 peptidase [M12.225] (38) | LVTATLGG (14) | (53) | |
| H–––- (1) | g | |||||
| Y–––- (5) | h | |||||
| Y–-IM– (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 813 | ADAMTS5 peptidase [M12.225] (38) | TATLGGEE (12) | (53) | |
| M–––- (3) | h | |||||
| –S––- (5) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 830 | ADAMTS5 peptidase [M12.225] (38) | PLPATVGV (16) | (53) | |
| I–––- (1) | h | |||||
| M–-I–- (2) | h | |||||
| -H––– (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 832 | ADAMTS5 peptidase [M12.225] (38) | PATVGVTQ (13) | (53) | |
| –-I–– (6) | h | |||||
| XX––– (1) | d | |||||
| Probable FKBP-type peptidyl-prolyl cis-trans isomerase 1, chloroplastic | Q9LM71 | 71 | Thylakoidal processing peptidase [S26.008] (52) | SSEARERR (4) | (49) | |
| –-G–– (1) | g | |||||
| XXXXXXXX (15) | a, d |
| Substrate . | UniProt accession . | P1 . | Peptidase [MEROPS ID] (total substrates) . | Replacements . | Possible cause . | Ref. . |
|---|---|---|---|---|---|---|
| Serine protease HTRA2, mitochondrial | O43464 | 211 | HtrA2 peptidase [S01.278] (56) | VRLLSGDT (5) | (37) | |
| –-P–– (4) | g | |||||
| Cytochrome C | P00022 | 1 | mitochondrial methionyl aminopeptidase [M24.028] (131) | MGDVE (35) | (38) | |
| -C–- (1) | g | |||||
| Coagulation factor XIII A chain | P00488 | 38 | thrombin [S01.217] (169) | VVPRGVNL (22) | (39) | |
| –-L–– (1) | g | |||||
| Insulin-1 | P01325 | 87 | proprotein convertase 2 [S08.073] (59) | RQKRGIVD (36) | (40) | |
| –WH-W-W (1) | a | |||||
| –A-X–R (1) | d | |||||
| Collagen alpha-2(I) chain | P02465 | 870 | cathepsin D [A01.009] (145) | APGFLGLP (15) | (41) | |
| –-I–– (21) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−1 [M10.001] (70) | GPQGLLGA (28) | (42) | |
| -T––– (8) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−8 [M10.002] (87) | GPQGLLGA (28) | (42) | |
| -T–P–- (8) | h | |||||
| Platelet-derived growth factor subunit A | P04085 | 86 | Furin [S08.071] (116) | RRKRSIEE (38) | (43) | |
| G-LT–– (1) | b | |||||
| L–X–– (1) | d | |||||
| Collagen alpha-2(IV) chain | P08572 | 1077 | kallikrein-related peptidase 14 [S01.029] (49) | APGRAGLY (6) | (44) | |
| –-S–– (7) | a | |||||
| –-L–– (5) | a | |||||
| –-A–– (2) | a | |||||
| –-I–– (1) | a, g | |||||
| –-V–– (3) | ||||||
| Collagen alpha-2(IV) chain | P08572 | 1109 | kallikrein-related peptidase 14 [S01.029] (49) | KGERGTTG (12) | (44) | |
| –QP-E– (7) | a | |||||
| ––-E– (2) | a | |||||
| –-L–– (2) | a | |||||
| –-V–– (1) | a | |||||
| Insulin-like growth factor-binding protein 1 | P08833 | 165 | Matriptase [S01.302] (26) | KALHVTNI (2) | (45) | |
| –D-N–- (1) | b | |||||
| -SXXXDD- (1) | d | |||||
| –V––- (2) | h | |||||
| –-E–D- (3) | h | |||||
| Acyl-CoA thioesterase I | P0ADA1 | 26 | Signal peptidase I [S26.001] (294) | RAAAADTL (19) | (46) | |
| X–––- (1) | d | |||||
| Protein ygiW | P0ADU5 | 20 | Signal peptidase I [S26.001] (294) | PVMAAEQG (10) | (47) | |
| ––-X– (1) | d | |||||
| Chymotrypsin inhibitor 3 | P10822 | 24 | Signalase (animal) 21 kDa component [S26.010) (363) | SSTADDDL (4) | (48) | |
| X–––- (7) | d | |||||
| –-M–– (1) | h | |||||
| Plastocyanin minor isoform, chloroplastic | P11490 | 72 | Thylakoidal processing peptidase [S26.008] (52) | NAMAMEVL (20) | (49) | |
| ––Q–- (2) | h | |||||
| –-D–– (1) | g | |||||
| 50S ribosomal protein L7Ae | P12743 | 1 | Methionyl aminopeptidase 2 [M24.002] (130) | MPVYV (2) | (50) | |
| KKMA–– (1) | d | |||||
| SKDK–– (8) | d | |||||
| MAR–- (1) | d | |||||
| Beta-crystallin B3 | P19141 | 4 | Calpain-1 [C02.001] (101) | MAEQHSTP (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| Beta-crystallin B3 | P19141 | 10 | Calpain-1 [C02.001] (101) | TPEQAAAG (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | P19174 | 770 | Caspase-7 [C14.004] (112) | AEPDYGAL (20) | (52) | |
| T–––- (1) | g | |||||
| Mimecan | P20774 | 219 | ADAMTS4 peptidase [M12.221] (57) | TFLYLDHN (26) | (53) | |
| -H––– (3) | h | |||||
| Mimecan | P20774 | 234 | ADAMTS4 peptidase [M12.221] (57) | NLPESLRV (23) | (53) | |
| X–––- (6) | d | |||||
| Trypsin inhibitor 2 | P26780 | 30 | Signalase (animal) 21 kDa component [S26.010] (363) | IKAQDSEC (7) | (54) | |
| –-H–– (2) | a | |||||
| 60S ribosomal protein L10 | P27635 | 180 | Granzyme B (Homo sapiens)-type) [S01.010] (348) | NADEFEDM (36) | (55) | |
| R–––- (2) | ||||||
| Chitinase 2 | P29027 | 22 | Signalase (animal) 21 kDa component [S26.010] (363) | GVQAAWSS (2) | (56) | |
| XX––– (1) | a, d | |||||
| Alpha-synuclein | P37840 | 122 | Calpain-1 | DPDNEAYE (34) | (57) | |
| [C02.001] (101) | –-D–– (1) | g | ||||
| –X––- (2) | b, d | |||||
| Cathepsin E | P43159 | 53 | Cathepsin E [A01.010] (64) | KVDMVQYT (14) | (35) | |
| -Y––– (11) | a | |||||
| -F––– (2) | h | |||||
| -H––– (3) | a | |||||
| –-G–– (2) | a | |||||
| –-T–– (2) | h | |||||
| ––H–- (5) | a | |||||
| –––H- (1) | g | |||||
| 40S ribosomal protein S25 | P62852 | 51 | Granzyme B, | LFDKATYD (9) | (55) | |
| rodent-type [S01.136] (231) | –XXXXX- (2) | b, d | ||||
| Hemoglobin subunit alpha | P69905 | 37 | Cathepsin D [A01.009] (145) | FLSFPTTK (42) | (34) | |
| ––-W– (1) | h | |||||
| Hemoglobin subunit alpha | P69905 | 109 | Cathepsin D [A01.009] (145) | LLVTLAAH (36) | (34) | |
| –––C- (6) | h | |||||
| Hemoglobin subunit alpha | P69905 | 110 | Cathepsin D [A01.009] (145) | LVTLAAHL (36) | (34) | |
| ––-C– (6) | h | |||||
| ABC transporter periplasmic-binding protein yphF | P77269 | 26 | Signal peptidase I [S26.001] (294) | FARAAEKE (22) | (47) | |
| –-T–– (1) | g | |||||
| Tyrosine-protein phosphatase non-receptor type 18 | Q61152 | 424 | Caspase-1 [C14.001] (60) | EVTDGAQT (4) | (58) | |
| –-G–– (4) | h | |||||
| ––R–- (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 810 | ADAMTS5 peptidase [M12.225] (38) | ALVTATLG (14) | (53) | |
| -H–-N– (1) | h | |||||
| ––-N– (2) | h | |||||
| –––M- (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 811 | ADAMTS5 peptidase [M12.225] (38) | LVTATLGG (14) | (53) | |
| H–––- (1) | g | |||||
| Y–––- (5) | h | |||||
| Y–-IM– (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 813 | ADAMTS5 peptidase [M12.225] (38) | TATLGGEE (12) | (53) | |
| M–––- (3) | h | |||||
| –S––- (5) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 830 | ADAMTS5 peptidase [M12.225] (38) | PLPATVGV (16) | (53) | |
| I–––- (1) | h | |||||
| M–-I–- (2) | h | |||||
| -H––– (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 832 | ADAMTS5 peptidase [M12.225] (38) | PATVGVTQ (13) | (53) | |
| –-I–– (6) | h | |||||
| XX––– (1) | d | |||||
| Probable FKBP-type peptidyl-prolyl cis-trans isomerase 1, chloroplastic | Q9LM71 | 71 | Thylakoidal processing peptidase [S26.008] (52) | SSEARERR (4) | (49) | |
| –-G–– (1) | g | |||||
| XXXXXXXX (15) | a, d |
The substrate name, UniProt accession, number of the residue occupying the P1 position in the known cleavage, the peptidase performing the cleavage (with MEROPS identifier in square brackets and the total of known substrates for the peptidase in parentheses), the sequence occupying P4–P4′ in the known cleavage and replacements unobserved in other substrates, the possible cause (a–h, see text for details), and the reference describing the cleavage are given. The numbers in parenthesis after the sequence are the number of homologues where the cleavage site is conserved (those identical to the known cleavage plus acceptable replacements) and the number of sequences where a replacement has occurred that has not been observed in any substrate for the peptidase. A hyphen indicates a conserved amino acid or an acceptable replacement, an ‘x’ indicates a gap character inserted in the alignment. A space indicates where no amino acid is possible (e.g. in P4, P3 and P2 for an aminopeptidase cleavage). Data are arranged by UniProt accession and the P1 position.
Assumed physiological cleavages that are not conserved in terms of peptidase substrate binding
| Substrate . | UniProt accession . | P1 . | Peptidase [MEROPS ID] (total substrates) . | Replacements . | Possible cause . | Ref. . |
|---|---|---|---|---|---|---|
| Serine protease HTRA2, mitochondrial | O43464 | 211 | HtrA2 peptidase [S01.278] (56) | VRLLSGDT (5) | (37) | |
| –-P–– (4) | g | |||||
| Cytochrome C | P00022 | 1 | mitochondrial methionyl aminopeptidase [M24.028] (131) | MGDVE (35) | (38) | |
| -C–- (1) | g | |||||
| Coagulation factor XIII A chain | P00488 | 38 | thrombin [S01.217] (169) | VVPRGVNL (22) | (39) | |
| –-L–– (1) | g | |||||
| Insulin-1 | P01325 | 87 | proprotein convertase 2 [S08.073] (59) | RQKRGIVD (36) | (40) | |
| –WH-W-W (1) | a | |||||
| –A-X–R (1) | d | |||||
| Collagen alpha-2(I) chain | P02465 | 870 | cathepsin D [A01.009] (145) | APGFLGLP (15) | (41) | |
| –-I–– (21) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−1 [M10.001] (70) | GPQGLLGA (28) | (42) | |
| -T––– (8) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−8 [M10.002] (87) | GPQGLLGA (28) | (42) | |
| -T–P–- (8) | h | |||||
| Platelet-derived growth factor subunit A | P04085 | 86 | Furin [S08.071] (116) | RRKRSIEE (38) | (43) | |
| G-LT–– (1) | b | |||||
| L–X–– (1) | d | |||||
| Collagen alpha-2(IV) chain | P08572 | 1077 | kallikrein-related peptidase 14 [S01.029] (49) | APGRAGLY (6) | (44) | |
| –-S–– (7) | a | |||||
| –-L–– (5) | a | |||||
| –-A–– (2) | a | |||||
| –-I–– (1) | a, g | |||||
| –-V–– (3) | ||||||
| Collagen alpha-2(IV) chain | P08572 | 1109 | kallikrein-related peptidase 14 [S01.029] (49) | KGERGTTG (12) | (44) | |
| –QP-E– (7) | a | |||||
| ––-E– (2) | a | |||||
| –-L–– (2) | a | |||||
| –-V–– (1) | a | |||||
| Insulin-like growth factor-binding protein 1 | P08833 | 165 | Matriptase [S01.302] (26) | KALHVTNI (2) | (45) | |
| –D-N–- (1) | b | |||||
| -SXXXDD- (1) | d | |||||
| –V––- (2) | h | |||||
| –-E–D- (3) | h | |||||
| Acyl-CoA thioesterase I | P0ADA1 | 26 | Signal peptidase I [S26.001] (294) | RAAAADTL (19) | (46) | |
| X–––- (1) | d | |||||
| Protein ygiW | P0ADU5 | 20 | Signal peptidase I [S26.001] (294) | PVMAAEQG (10) | (47) | |
| ––-X– (1) | d | |||||
| Chymotrypsin inhibitor 3 | P10822 | 24 | Signalase (animal) 21 kDa component [S26.010) (363) | SSTADDDL (4) | (48) | |
| X–––- (7) | d | |||||
| –-M–– (1) | h | |||||
| Plastocyanin minor isoform, chloroplastic | P11490 | 72 | Thylakoidal processing peptidase [S26.008] (52) | NAMAMEVL (20) | (49) | |
| ––Q–- (2) | h | |||||
| –-D–– (1) | g | |||||
| 50S ribosomal protein L7Ae | P12743 | 1 | Methionyl aminopeptidase 2 [M24.002] (130) | MPVYV (2) | (50) | |
| KKMA–– (1) | d | |||||
| SKDK–– (8) | d | |||||
| MAR–- (1) | d | |||||
| Beta-crystallin B3 | P19141 | 4 | Calpain-1 [C02.001] (101) | MAEQHSTP (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| Beta-crystallin B3 | P19141 | 10 | Calpain-1 [C02.001] (101) | TPEQAAAG (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | P19174 | 770 | Caspase-7 [C14.004] (112) | AEPDYGAL (20) | (52) | |
| T–––- (1) | g | |||||
| Mimecan | P20774 | 219 | ADAMTS4 peptidase [M12.221] (57) | TFLYLDHN (26) | (53) | |
| -H––– (3) | h | |||||
| Mimecan | P20774 | 234 | ADAMTS4 peptidase [M12.221] (57) | NLPESLRV (23) | (53) | |
| X–––- (6) | d | |||||
| Trypsin inhibitor 2 | P26780 | 30 | Signalase (animal) 21 kDa component [S26.010] (363) | IKAQDSEC (7) | (54) | |
| –-H–– (2) | a | |||||
| 60S ribosomal protein L10 | P27635 | 180 | Granzyme B (Homo sapiens)-type) [S01.010] (348) | NADEFEDM (36) | (55) | |
| R–––- (2) | ||||||
| Chitinase 2 | P29027 | 22 | Signalase (animal) 21 kDa component [S26.010] (363) | GVQAAWSS (2) | (56) | |
| XX––– (1) | a, d | |||||
| Alpha-synuclein | P37840 | 122 | Calpain-1 | DPDNEAYE (34) | (57) | |
| [C02.001] (101) | –-D–– (1) | g | ||||
| –X––- (2) | b, d | |||||
| Cathepsin E | P43159 | 53 | Cathepsin E [A01.010] (64) | KVDMVQYT (14) | (35) | |
| -Y––– (11) | a | |||||
| -F––– (2) | h | |||||
| -H––– (3) | a | |||||
| –-G–– (2) | a | |||||
| –-T–– (2) | h | |||||
| ––H–- (5) | a | |||||
| –––H- (1) | g | |||||
| 40S ribosomal protein S25 | P62852 | 51 | Granzyme B, | LFDKATYD (9) | (55) | |
| rodent-type [S01.136] (231) | –XXXXX- (2) | b, d | ||||
| Hemoglobin subunit alpha | P69905 | 37 | Cathepsin D [A01.009] (145) | FLSFPTTK (42) | (34) | |
| ––-W– (1) | h | |||||
| Hemoglobin subunit alpha | P69905 | 109 | Cathepsin D [A01.009] (145) | LLVTLAAH (36) | (34) | |
| –––C- (6) | h | |||||
| Hemoglobin subunit alpha | P69905 | 110 | Cathepsin D [A01.009] (145) | LVTLAAHL (36) | (34) | |
| ––-C– (6) | h | |||||
| ABC transporter periplasmic-binding protein yphF | P77269 | 26 | Signal peptidase I [S26.001] (294) | FARAAEKE (22) | (47) | |
| –-T–– (1) | g | |||||
| Tyrosine-protein phosphatase non-receptor type 18 | Q61152 | 424 | Caspase-1 [C14.001] (60) | EVTDGAQT (4) | (58) | |
| –-G–– (4) | h | |||||
| ––R–- (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 810 | ADAMTS5 peptidase [M12.225] (38) | ALVTATLG (14) | (53) | |
| -H–-N– (1) | h | |||||
| ––-N– (2) | h | |||||
| –––M- (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 811 | ADAMTS5 peptidase [M12.225] (38) | LVTATLGG (14) | (53) | |
| H–––- (1) | g | |||||
| Y–––- (5) | h | |||||
| Y–-IM– (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 813 | ADAMTS5 peptidase [M12.225] (38) | TATLGGEE (12) | (53) | |
| M–––- (3) | h | |||||
| –S––- (5) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 830 | ADAMTS5 peptidase [M12.225] (38) | PLPATVGV (16) | (53) | |
| I–––- (1) | h | |||||
| M–-I–- (2) | h | |||||
| -H––– (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 832 | ADAMTS5 peptidase [M12.225] (38) | PATVGVTQ (13) | (53) | |
| –-I–– (6) | h | |||||
| XX––– (1) | d | |||||
| Probable FKBP-type peptidyl-prolyl cis-trans isomerase 1, chloroplastic | Q9LM71 | 71 | Thylakoidal processing peptidase [S26.008] (52) | SSEARERR (4) | (49) | |
| –-G–– (1) | g | |||||
| XXXXXXXX (15) | a, d |
| Substrate . | UniProt accession . | P1 . | Peptidase [MEROPS ID] (total substrates) . | Replacements . | Possible cause . | Ref. . |
|---|---|---|---|---|---|---|
| Serine protease HTRA2, mitochondrial | O43464 | 211 | HtrA2 peptidase [S01.278] (56) | VRLLSGDT (5) | (37) | |
| –-P–– (4) | g | |||||
| Cytochrome C | P00022 | 1 | mitochondrial methionyl aminopeptidase [M24.028] (131) | MGDVE (35) | (38) | |
| -C–- (1) | g | |||||
| Coagulation factor XIII A chain | P00488 | 38 | thrombin [S01.217] (169) | VVPRGVNL (22) | (39) | |
| –-L–– (1) | g | |||||
| Insulin-1 | P01325 | 87 | proprotein convertase 2 [S08.073] (59) | RQKRGIVD (36) | (40) | |
| –WH-W-W (1) | a | |||||
| –A-X–R (1) | d | |||||
| Collagen alpha-2(I) chain | P02465 | 870 | cathepsin D [A01.009] (145) | APGFLGLP (15) | (41) | |
| –-I–– (21) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−1 [M10.001] (70) | GPQGLLGA (28) | (42) | |
| -T––– (8) | h | |||||
| Collagen alpha-2(I) chain | P02465 | 863 | matrix metallopeptidase−8 [M10.002] (87) | GPQGLLGA (28) | (42) | |
| -T–P–- (8) | h | |||||
| Platelet-derived growth factor subunit A | P04085 | 86 | Furin [S08.071] (116) | RRKRSIEE (38) | (43) | |
| G-LT–– (1) | b | |||||
| L–X–– (1) | d | |||||
| Collagen alpha-2(IV) chain | P08572 | 1077 | kallikrein-related peptidase 14 [S01.029] (49) | APGRAGLY (6) | (44) | |
| –-S–– (7) | a | |||||
| –-L–– (5) | a | |||||
| –-A–– (2) | a | |||||
| –-I–– (1) | a, g | |||||
| –-V–– (3) | ||||||
| Collagen alpha-2(IV) chain | P08572 | 1109 | kallikrein-related peptidase 14 [S01.029] (49) | KGERGTTG (12) | (44) | |
| –QP-E– (7) | a | |||||
| ––-E– (2) | a | |||||
| –-L–– (2) | a | |||||
| –-V–– (1) | a | |||||
| Insulin-like growth factor-binding protein 1 | P08833 | 165 | Matriptase [S01.302] (26) | KALHVTNI (2) | (45) | |
| –D-N–- (1) | b | |||||
| -SXXXDD- (1) | d | |||||
| –V––- (2) | h | |||||
| –-E–D- (3) | h | |||||
| Acyl-CoA thioesterase I | P0ADA1 | 26 | Signal peptidase I [S26.001] (294) | RAAAADTL (19) | (46) | |
| X–––- (1) | d | |||||
| Protein ygiW | P0ADU5 | 20 | Signal peptidase I [S26.001] (294) | PVMAAEQG (10) | (47) | |
| ––-X– (1) | d | |||||
| Chymotrypsin inhibitor 3 | P10822 | 24 | Signalase (animal) 21 kDa component [S26.010) (363) | SSTADDDL (4) | (48) | |
| X–––- (7) | d | |||||
| –-M–– (1) | h | |||||
| Plastocyanin minor isoform, chloroplastic | P11490 | 72 | Thylakoidal processing peptidase [S26.008] (52) | NAMAMEVL (20) | (49) | |
| ––Q–- (2) | h | |||||
| –-D–– (1) | g | |||||
| 50S ribosomal protein L7Ae | P12743 | 1 | Methionyl aminopeptidase 2 [M24.002] (130) | MPVYV (2) | (50) | |
| KKMA–– (1) | d | |||||
| SKDK–– (8) | d | |||||
| MAR–- (1) | d | |||||
| Beta-crystallin B3 | P19141 | 4 | Calpain-1 [C02.001] (101) | MAEQHSTP (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| Beta-crystallin B3 | P19141 | 10 | Calpain-1 [C02.001] (101) | TPEQAAAG (10) | (51) | |
| XXXXXXXX (23) | a, d | |||||
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | P19174 | 770 | Caspase-7 [C14.004] (112) | AEPDYGAL (20) | (52) | |
| T–––- (1) | g | |||||
| Mimecan | P20774 | 219 | ADAMTS4 peptidase [M12.221] (57) | TFLYLDHN (26) | (53) | |
| -H––– (3) | h | |||||
| Mimecan | P20774 | 234 | ADAMTS4 peptidase [M12.221] (57) | NLPESLRV (23) | (53) | |
| X–––- (6) | d | |||||
| Trypsin inhibitor 2 | P26780 | 30 | Signalase (animal) 21 kDa component [S26.010] (363) | IKAQDSEC (7) | (54) | |
| –-H–– (2) | a | |||||
| 60S ribosomal protein L10 | P27635 | 180 | Granzyme B (Homo sapiens)-type) [S01.010] (348) | NADEFEDM (36) | (55) | |
| R–––- (2) | ||||||
| Chitinase 2 | P29027 | 22 | Signalase (animal) 21 kDa component [S26.010] (363) | GVQAAWSS (2) | (56) | |
| XX––– (1) | a, d | |||||
| Alpha-synuclein | P37840 | 122 | Calpain-1 | DPDNEAYE (34) | (57) | |
| [C02.001] (101) | –-D–– (1) | g | ||||
| –X––- (2) | b, d | |||||
| Cathepsin E | P43159 | 53 | Cathepsin E [A01.010] (64) | KVDMVQYT (14) | (35) | |
| -Y––– (11) | a | |||||
| -F––– (2) | h | |||||
| -H––– (3) | a | |||||
| –-G–– (2) | a | |||||
| –-T–– (2) | h | |||||
| ––H–- (5) | a | |||||
| –––H- (1) | g | |||||
| 40S ribosomal protein S25 | P62852 | 51 | Granzyme B, | LFDKATYD (9) | (55) | |
| rodent-type [S01.136] (231) | –XXXXX- (2) | b, d | ||||
| Hemoglobin subunit alpha | P69905 | 37 | Cathepsin D [A01.009] (145) | FLSFPTTK (42) | (34) | |
| ––-W– (1) | h | |||||
| Hemoglobin subunit alpha | P69905 | 109 | Cathepsin D [A01.009] (145) | LLVTLAAH (36) | (34) | |
| –––C- (6) | h | |||||
| Hemoglobin subunit alpha | P69905 | 110 | Cathepsin D [A01.009] (145) | LVTLAAHL (36) | (34) | |
| ––-C– (6) | h | |||||
| ABC transporter periplasmic-binding protein yphF | P77269 | 26 | Signal peptidase I [S26.001] (294) | FARAAEKE (22) | (47) | |
| –-T–– (1) | g | |||||
| Tyrosine-protein phosphatase non-receptor type 18 | Q61152 | 424 | Caspase-1 [C14.001] (60) | EVTDGAQT (4) | (58) | |
| –-G–– (4) | h | |||||
| ––R–- (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 810 | ADAMTS5 peptidase [M12.225] (38) | ALVTATLG (14) | (53) | |
| -H–-N– (1) | h | |||||
| ––-N– (2) | h | |||||
| –––M- (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 811 | ADAMTS5 peptidase [M12.225] (38) | LVTATLGG (14) | (53) | |
| H–––- (1) | g | |||||
| Y–––- (5) | h | |||||
| Y–-IM– (2) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 813 | ADAMTS5 peptidase [M12.225] (38) | TATLGGEE (12) | (53) | |
| M–––- (3) | h | |||||
| –S––- (5) | h | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 830 | ADAMTS5 peptidase [M12.225] (38) | PLPATVGV (16) | (53) | |
| I–––- (1) | h | |||||
| M–-I–- (2) | h | |||||
| -H––– (1) | g | |||||
| Cartilage intermediate layer protein 2 | Q8IUL8 | 832 | ADAMTS5 peptidase [M12.225] (38) | PATVGVTQ (13) | (53) | |
| –-I–– (6) | h | |||||
| XX––– (1) | d | |||||
| Probable FKBP-type peptidyl-prolyl cis-trans isomerase 1, chloroplastic | Q9LM71 | 71 | Thylakoidal processing peptidase [S26.008] (52) | SSEARERR (4) | (49) | |
| –-G–– (1) | g | |||||
| XXXXXXXX (15) | a, d |
The substrate name, UniProt accession, number of the residue occupying the P1 position in the known cleavage, the peptidase performing the cleavage (with MEROPS identifier in square brackets and the total of known substrates for the peptidase in parentheses), the sequence occupying P4–P4′ in the known cleavage and replacements unobserved in other substrates, the possible cause (a–h, see text for details), and the reference describing the cleavage are given. The numbers in parenthesis after the sequence are the number of homologues where the cleavage site is conserved (those identical to the known cleavage plus acceptable replacements) and the number of sequences where a replacement has occurred that has not been observed in any substrate for the peptidase. A hyphen indicates a conserved amino acid or an acceptable replacement, an ‘x’ indicates a gap character inserted in the alignment. A space indicates where no amino acid is possible (e.g. in P4, P3 and P2 for an aminopeptidase cleavage). Data are arranged by UniProt accession and the P1 position.
There are a number of possible causes for a cleavage site not to be conserved which are listed below.
The UniRef50 entry might include paralogous sequences which although at least 50% identical to the sequence with the known cleavage, might be processed or degraded differently and there is no evolutionary pressure to maintain the known cleavage site. Where a cleavage site was not conserved, a paralogue was identified in an alignment as a second protein from the same species that was clearly not a splice variant.
UniRef50 entries contain many translated genes from genome sequencing projects; gene finding in eukaryote genomes is notoriously difficult and it is possible that erroneous gene building has resulted, for example, in the loss of the exon encoding the cleavage site or the inclusion of part of an intron in its place.
It is also probable that for some peptidases there are not enough substrates known to be sure that any amino acid is really excluded from a particular binding site. The number of substrates known for each peptidase is included in Table 4, because the greater the number of substrates the more likely that an amino acid is really atypical and not just unobserved.
The alignment is incorrect. This is unlikely given the close relationship between the sequences, which are all 50% or more identical; however there are situations where an insert or deletion occurs within the range P4–P4′.
Some endogenous cleavages (for example removal of signal and transit peptides) may be the result of more than one cleavage, because aminopeptidases nibble away the N-terminus (1), and may thus be incorrectly mapped to the specificity of the leader peptidase.
It is theoretically possible that if the substrate and peptidase are from the same organism both will have evolved to accommodate a change in the cleavage position.
A single residue mismatch may also be due to a single-base sequencing error. Potential errors of this kind can be identified using a codon dictionary, provided the atypical residue could be the result of a single base change, and that it is the only residue not conserved, regardless of the number of sequences in the alignment.
Some cleavages regarded as ‘physiological’ are actually fortuitous. If a cleavage site is extremely poorly conserved it is unlikely to be physiologically relevant.
Where it is possible to suggest a cause why a cleavage site is not conserved this is indicated in Table 4 by the letters a–h. Included in category d, where insertions and or deletions occur in the homologous cleavage sites, is 50S ribosomal protein L7Ae (UniProt accession P12743). There are N-terminal extensions to most homologues so that the known methionyl aminopeptidase 2-cleavage site is not aligned. Five of these sequences may be derived from erroneous gene builds (point b). The UniRef50 database entry for 60S ribosomal protein L10 (P27635) includes a wide range of species (the cleavage is known in the human protein) and the peptidase performing the cleavage (granzyme B) is not present in Paracoccidiodes brasiliensis, where the substrate cleavage is also not conserved. The replacements that are reported as atypical in hemoglobin subunit alpha (P69905) by Schistosoma cathepsin D (A01.009) (34) are the rarest naturally occurring amino acids, tryptophan and cysteine, and despite there being 109 known cleavages for this peptidase, this may still not be enough to properly exclude these rare amino acids. On the other hand, this is the cleavage of a host protein by a parasite peptidase and the specificity may have adapted to limit the availability of hosts.
None of the cleavages listed in Table 4 has been assigned to cause f above, namely where changes in the substrate cleavage site may be mirrored by changes in peptidase specificity. Without modelling the substrate binding sites, if that were possible, detecting this situation is difficult. However, the autolytic processing of cathepsin E (P43159) may be such an example (35).
In some cases, a poorly conserved cleavage site may represent a pathological condition in the species where the cleavage was first identified. For example, despite there being few cleavages for cathepsin H, the reported cleavages in the BID protein (36) are in particularly poorly conserved regions (see Figure 3). Cleavage of the BID protein leads to the induction of apoptosis. That the cleavage sites are not well conserved amongst mammalian orthologues is not surprising given that the cytoplasmic substrate and the lysosomal peptidase should not meet under normal circumstances. The mouse protein in which the cleavage was identified may therefore be unusually susceptible to cleavage should the lysosomal membrane be ruptured.
The specificity logos and frequency matrices for all peptidases with 10 or more known substrate cleavages are already available in the MEROPS database. Alignments are also available for all protein substrates that have a corresponding UniRef50 entry, showing conservation of both physiological and non-physiological cleavages. The next release of the database will include tables showing comparative peptidase specificity in terms of preference for both amino acid and amino acid type.
Conclusions
The MEROPS database includes over 39 000 cleavages in substrates (synthetic and naturally occurring) which have been collected from the literature. These are classified as physiological or non-physiological, depending on whether the substrate is naturally occurring and if it is in native conformation. At least one substrate is known for 45% of the different peptidases identified in the MEROPS database. Displays in the database give insights into peptidase specificity and to the conservation of cleavage sites amongst orthologous proteins. The data provide a substantial training set for algorithms to predict peptidase substrates and cleavage positions in those substrates. The data may also be useful for the design of inhibitors and engineering novel specificities into peptidases.
By examining the conservation of cleavage sites in protein substrates in terms of peptidase substrate binding sites, it is clear that there are a number of cleavages where atypical replacements occur. Many of these can be explained by gene build or sequencing errors, inserts or deletions in the region around the cleavage site, or the alignments contain one or more paralogues in which cleavage may be absent or different. In a few cases it is possible that more than one peptidase is involved in processing, or there may not be enough known substrates for some peptidases to be sure that an atypical replacement is really unacceptable. A number of substrate cleavages that may be fortuitous and not of any physiological relevance have been identified.
This cleavage set is freely available and can be downloaded from the MEROPS FTP site (ftp://ftp.sanger.ac.uk/pub/MEROPS/current_release/database_files/Substrate_search.txt).
Funding
Wellcome Trust [grant number WT077044/Z/05/Z]. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Acknowledgements
the author would like to thank Dr Alan Barrett, Chai Yin Kok, Jun Kong, Matias Piipari, Matthew Jenner and Olivia Harris for helping to collect and/or enter cleavage data, Jun Kong for devising the specificity logo software, Dr Penelope Coggill for reading the manuscript and Dr Alex Bateman for guidance and useful discussions.



