-
PDF
- Split View
-
Views
-
Cite
Cite
Laurens G. Wilming, Elizabeth A. Hart, Penny C. Coggill, Roger Horton, James G. R. Gilbert, Chris Clee, Matt Jones, Christine Lloyd, Sophie Palmer, Sarah Sims, Siobhan Whitehead, David Wiley, Stephan Beck, Jennifer L. Harrow, Sequencing and comparative analysis of the gorilla MHC genomic sequence, Database, Volume 2013, 2013, bat011, https://doi.org/10.1093/database/bat011
Close - Share Icon Share
Abstract
Major histocompatibility complex (MHC) genes play a critical role in vertebrate immune response and because the MHC is linked to a significant number of auto-immune and other diseases it is of great medical interest. Here we describe the clone-based sequencing and subsequent annotation of the MHC region of the gorilla genome. Because the MHC is subject to extensive variation, both structural and sequence-wise, it is not readily amenable to study in whole genome shotgun sequence such as the recently published gorilla genome. The variation of the MHC also makes it of evolutionary interest and therefore we analyse the sequence in the context of human and chimpanzee. In our comparisons with human and re-annotated chimpanzee MHC sequence we find that gorilla has a trimodular RCCX cluster, versus the reference human bimodular cluster, and additional copies of Class I (pseudo)genes between Gogo-K and Gogo-A (the orthologues of HLA-K and -A). We also find that Gogo-H (and Patr-H) is coding versus the HLA-H pseudogene and, conversely, there is a Gogo-DQB2 pseudogene versus the HLA-DQB2 coding gene. Our analysis, which is freely available through the VEGA genome browser, provides the research community with a comprehensive dataset for comparative and evolutionary research of the MHC.
Introduction
The major histocompatibility complex, MHC, is the multi-gene region of the genome crucial for the vertebrate immune response. It is the most gene-dense and polymorphic region of the mammalian genome (1, 2). In conjunction with those of the leucocyte receptor complex, its cell surface–expressed products are responsible for transplant effectiveness, reproductive success, autoimmunity and much of the resistance to infectious diseases. Recent studies on eight human haplotypes have provided a detailed picture of the haplotype structure and gene polymorphism of this region (3, 4). In addition, the Wellcome Trust Case Control Consortium genome-wide association study for seven common diseases (5) has found the highest associations for two autoimmune diseases—type I diabetes and rheumatoid arthritis—to be with the MHC. These combined findings offer, for the first time, a secure baseline from which to start evaluating the significance of the evolutionary development of the MHC as a whole.
The MHC region is divided into sub-regions according to the type or function of the genes they contain: extended class I, classical class I, classical class III, classical class II and extended class II (6). The extended class I region contains, for example, histone and olfactory receptor genes, the classical class III region contains the complement factor genes, and the primary role of the classical MHC class I and II genes is to encode key receptor molecules that recognise foreign peptides and present them to specialist immune cells to initiate an immune response. Each MHC molecule comprises a non-variable immunoglobulin ‘stalk’ that anchors the molecule to the cell surface and a basket-like receptor that forms the peptide-binding region. It is these peptide-binding regions that are highly polymorphic in all primates (7). Comparing the polymorphism in other great apes with that in humans may indicate how evolution is being driven. Analysis of rhesus macaque MHC genes has informed the study of the region as a whole, but the human genome, in general, is more similar to the chimpanzee genome, with which it shares 98.8% nucleotide and >99% amino acid identity (8). However, de novo or species-specific polymorphisms were implied by diversity centred largely on the HLA-A and HLA-B/C clusters (9). There are also a few specific gene regions where the similarity to gorilla is higher (10), so the study of the gorilla as an out-group between human and chimpanzee will add another layer to the understanding of the evolutionary changes that have occurred in humans in their ability to combat infection and mount appropriate immune responses.
In the gorilla, the MHC cluster is located on the equivalent to human chromosome 6 (where the human MHC gene cluster is located); depending on the nomenclature used, this is chromosome 5 (when numbering the chromosomes 1–23, where chromosomes 11 and 12 are equivalent to q and p arms of human chromosome 2, respectively) or chromosome 6 (when numbering the human chromosome 2 equivalents as 2p and 2q or 2a and 2b) (11–13). The bacterial artificial chromosome (BAC) library containing the region was made from a heterozygous gorilla and we have attempted to give contiguous single haplotype coverage of the whole 4-Mb region that includes MHC class I, III and II. But it is clear that, at least across the class II—DRB—region, the two haplotypes present are distinct. Here we provide the first complete reference sequence and gene map of the full MHC region in the gorilla. As this sequence is based on contiguous BAC clones, as opposed to whole-genome shotgun assembly used for the genome of female gorilla ‘Kamilah' (14), we are able to provide comprehensive annotation of this region. We make direct comparisons with the reference human genome and the chimpanzee MHC sequence (9), both of which have been annotated by the HAVANA group and published in the Vertebrate Genome Annotation (VEGA) genome browser (15). Similarities and differences between orthologous genes in other primates are discussed.
Results and discussion
Gorilla MHC gene map
The 29 BAC clones that comprise the gorilla MHC span a region of 4.64-Mb and are listed in Table 1, together with the variations found within the 2000 base-pair or longer overlaps between clones. A map of the entire region is shown in Figure 1. Within this contiguous region, 305 gene loci have been identified and annotated: 155 of these are known genes that are predicted to be functional, 5 loci are classified as novel CDS (see ‘Material and Methods: Sequence annotation’ section for definitions), 15 are known transcripts, 5 are novel transcripts and 20 are putative transcripts. The remaining 105 loci have been classified as pseudogenes: 44 as processed pseudogenes, 59 as unprocessed pseudogenes, 1 as a polymorphic pseudogene and 1 as a transcribed pseudogene. The full annotation is available online at the VEGA database (15, 16). It should be noted that virtually all gorilla transcript models have been built on the basis of homology support by human transcripts or proteins. Notable exceptions are the following transcripts, which are supported by native locus-specific mRNAs: variant 001 of TAP1 (TAP1-001), TAP2-001, Gogo-A-006, Gogo-C-001 and Gogo-DMB-001.

Feature map of the gorilla MHC, modified from the VEGA browser (release 50, December 2012). Each locus is labelled with a name, coloured according to type (see legend at bottom) and with indication of orientation (angle bracket before or after the name) and position within the region. The tiling path of the sequenced BACs is shown at the top of each panel (labelled contigs), with clones in alternating dark and light blue and, space permitting, with accession numbers. At the top and bottom of each panel, a size scale is shown. The regions highlighted in Figure 2 are marked with green bars at the bottom of a panel and labelled with the figure section identifier.
The clone names, versioned accession numbers and variation between the BACs that make up the MHC of gorilla ‘Frank’
| Clone name . | ENA accession . | Number of . | Overlap length (bp) . | ||
|---|---|---|---|---|---|
| . | . | SNPs . | Deletions . | Insertions . | . |
| CH255-522B23 | CU104671.1 | 0 | 0 | 0 | 2000 |
| CH255-127C23 | CU104653.1 | 0 | 0 | 0 | 2000 |
| CH255-37P17 | CU104661.1 | 0 | 0 | 0 | 2000 |
| CH255-179G12 | CT025620.2 | 0 | 0 | 0 | 2000 |
| CH255-451A14 | CU104667.1 | 0 | 0 | 0 | 2000 |
| CH255-405H12 | CU104665.1 | 0 | 0 | 0 | 2000 |
| CH255-259J17 | CU104658.1 | 0 | 0 | 0 | 2000 |
| CH255-39I5 | CU104664.1 | 0 | 0 | 0 | 2000 |
| CH255-83O18 | CU104675.1 | 0 | 0 | 0 | 2000 |
| CH255-289P22 | CU104659.1 | 0 | 0 | 0 | 2000 |
| CH255-201E9 | CU104656.1 | 0 | 0 | 0 | 2000 |
| CH255-478L19 | CU104669.1 | 0 | 0 | 0 | 2000 |
| CH255-48E14 | CU104670.1 | 0 | 0 | 0 | 65339 |
| CH255-386C2 | CU104662.1 | 0 | 0 | 0 | 2000 |
| CH255-375N4 | CU104660.1 | 0 | 0 | 0 | 37060 |
| CH255-13G2 | CU104654.2 | 8 | 4 | 3 | 24914 |
| CH255-415I16 | CU104666.2 | 98 | 20 | 22 | 103353 |
| CH255-559J12 | CU104673.1 | 0 | 0 | 0 | 2000 |
| CH255-397I3 | CU104663.1 | 0 | 0 | 0 | 2000 |
| CH255-469C9 | CU104668.1 | 0 | 0 | 0 | 2635 |
| CH255-56N15 | CU104674.1 | 4 | 2 | 1 | 32416 |
| CH255-114D6 | CU104652.1 | 0 | 0 | 0 | 2000 |
| CH255-58L21 | CU104676.1 | 0 | 0 | 0 | 2000 |
| CH255-351B13 | CT025711.1 | 0 | 0 | 0 | 29819 |
| CH255-354J20 | CT025621.2 | 0 | 0 | 0 | 2000 |
| CH255-336G22 | CT025558.1 | 0 | 0 | 0 | 2000 |
| CH255-191J6 | CU104655.1 | 0 | 0 | 0 | 2000 |
| CH255-206J13 | CU104657.1 | 101 | 14 | 16 | 59844 |
| CH255-529K7 | CU104672.1 | ||||
| Clone name . | ENA accession . | Number of . | Overlap length (bp) . | ||
|---|---|---|---|---|---|
| . | . | SNPs . | Deletions . | Insertions . | . |
| CH255-522B23 | CU104671.1 | 0 | 0 | 0 | 2000 |
| CH255-127C23 | CU104653.1 | 0 | 0 | 0 | 2000 |
| CH255-37P17 | CU104661.1 | 0 | 0 | 0 | 2000 |
| CH255-179G12 | CT025620.2 | 0 | 0 | 0 | 2000 |
| CH255-451A14 | CU104667.1 | 0 | 0 | 0 | 2000 |
| CH255-405H12 | CU104665.1 | 0 | 0 | 0 | 2000 |
| CH255-259J17 | CU104658.1 | 0 | 0 | 0 | 2000 |
| CH255-39I5 | CU104664.1 | 0 | 0 | 0 | 2000 |
| CH255-83O18 | CU104675.1 | 0 | 0 | 0 | 2000 |
| CH255-289P22 | CU104659.1 | 0 | 0 | 0 | 2000 |
| CH255-201E9 | CU104656.1 | 0 | 0 | 0 | 2000 |
| CH255-478L19 | CU104669.1 | 0 | 0 | 0 | 2000 |
| CH255-48E14 | CU104670.1 | 0 | 0 | 0 | 65339 |
| CH255-386C2 | CU104662.1 | 0 | 0 | 0 | 2000 |
| CH255-375N4 | CU104660.1 | 0 | 0 | 0 | 37060 |
| CH255-13G2 | CU104654.2 | 8 | 4 | 3 | 24914 |
| CH255-415I16 | CU104666.2 | 98 | 20 | 22 | 103353 |
| CH255-559J12 | CU104673.1 | 0 | 0 | 0 | 2000 |
| CH255-397I3 | CU104663.1 | 0 | 0 | 0 | 2000 |
| CH255-469C9 | CU104668.1 | 0 | 0 | 0 | 2635 |
| CH255-56N15 | CU104674.1 | 4 | 2 | 1 | 32416 |
| CH255-114D6 | CU104652.1 | 0 | 0 | 0 | 2000 |
| CH255-58L21 | CU104676.1 | 0 | 0 | 0 | 2000 |
| CH255-351B13 | CT025711.1 | 0 | 0 | 0 | 29819 |
| CH255-354J20 | CT025621.2 | 0 | 0 | 0 | 2000 |
| CH255-336G22 | CT025558.1 | 0 | 0 | 0 | 2000 |
| CH255-191J6 | CU104655.1 | 0 | 0 | 0 | 2000 |
| CH255-206J13 | CU104657.1 | 101 | 14 | 16 | 59844 |
| CH255-529K7 | CU104672.1 | ||||
Variation shown is between the clone listed on that row and the next, in the length of the overlap between the two. Clones are listed in the order of contiguous overlap.
SNPs = single nucleotide polymorphisms.
The clone names, versioned accession numbers and variation between the BACs that make up the MHC of gorilla ‘Frank’
| Clone name . | ENA accession . | Number of . | Overlap length (bp) . | ||
|---|---|---|---|---|---|
| . | . | SNPs . | Deletions . | Insertions . | . |
| CH255-522B23 | CU104671.1 | 0 | 0 | 0 | 2000 |
| CH255-127C23 | CU104653.1 | 0 | 0 | 0 | 2000 |
| CH255-37P17 | CU104661.1 | 0 | 0 | 0 | 2000 |
| CH255-179G12 | CT025620.2 | 0 | 0 | 0 | 2000 |
| CH255-451A14 | CU104667.1 | 0 | 0 | 0 | 2000 |
| CH255-405H12 | CU104665.1 | 0 | 0 | 0 | 2000 |
| CH255-259J17 | CU104658.1 | 0 | 0 | 0 | 2000 |
| CH255-39I5 | CU104664.1 | 0 | 0 | 0 | 2000 |
| CH255-83O18 | CU104675.1 | 0 | 0 | 0 | 2000 |
| CH255-289P22 | CU104659.1 | 0 | 0 | 0 | 2000 |
| CH255-201E9 | CU104656.1 | 0 | 0 | 0 | 2000 |
| CH255-478L19 | CU104669.1 | 0 | 0 | 0 | 2000 |
| CH255-48E14 | CU104670.1 | 0 | 0 | 0 | 65339 |
| CH255-386C2 | CU104662.1 | 0 | 0 | 0 | 2000 |
| CH255-375N4 | CU104660.1 | 0 | 0 | 0 | 37060 |
| CH255-13G2 | CU104654.2 | 8 | 4 | 3 | 24914 |
| CH255-415I16 | CU104666.2 | 98 | 20 | 22 | 103353 |
| CH255-559J12 | CU104673.1 | 0 | 0 | 0 | 2000 |
| CH255-397I3 | CU104663.1 | 0 | 0 | 0 | 2000 |
| CH255-469C9 | CU104668.1 | 0 | 0 | 0 | 2635 |
| CH255-56N15 | CU104674.1 | 4 | 2 | 1 | 32416 |
| CH255-114D6 | CU104652.1 | 0 | 0 | 0 | 2000 |
| CH255-58L21 | CU104676.1 | 0 | 0 | 0 | 2000 |
| CH255-351B13 | CT025711.1 | 0 | 0 | 0 | 29819 |
| CH255-354J20 | CT025621.2 | 0 | 0 | 0 | 2000 |
| CH255-336G22 | CT025558.1 | 0 | 0 | 0 | 2000 |
| CH255-191J6 | CU104655.1 | 0 | 0 | 0 | 2000 |
| CH255-206J13 | CU104657.1 | 101 | 14 | 16 | 59844 |
| CH255-529K7 | CU104672.1 | ||||
| Clone name . | ENA accession . | Number of . | Overlap length (bp) . | ||
|---|---|---|---|---|---|
| . | . | SNPs . | Deletions . | Insertions . | . |
| CH255-522B23 | CU104671.1 | 0 | 0 | 0 | 2000 |
| CH255-127C23 | CU104653.1 | 0 | 0 | 0 | 2000 |
| CH255-37P17 | CU104661.1 | 0 | 0 | 0 | 2000 |
| CH255-179G12 | CT025620.2 | 0 | 0 | 0 | 2000 |
| CH255-451A14 | CU104667.1 | 0 | 0 | 0 | 2000 |
| CH255-405H12 | CU104665.1 | 0 | 0 | 0 | 2000 |
| CH255-259J17 | CU104658.1 | 0 | 0 | 0 | 2000 |
| CH255-39I5 | CU104664.1 | 0 | 0 | 0 | 2000 |
| CH255-83O18 | CU104675.1 | 0 | 0 | 0 | 2000 |
| CH255-289P22 | CU104659.1 | 0 | 0 | 0 | 2000 |
| CH255-201E9 | CU104656.1 | 0 | 0 | 0 | 2000 |
| CH255-478L19 | CU104669.1 | 0 | 0 | 0 | 2000 |
| CH255-48E14 | CU104670.1 | 0 | 0 | 0 | 65339 |
| CH255-386C2 | CU104662.1 | 0 | 0 | 0 | 2000 |
| CH255-375N4 | CU104660.1 | 0 | 0 | 0 | 37060 |
| CH255-13G2 | CU104654.2 | 8 | 4 | 3 | 24914 |
| CH255-415I16 | CU104666.2 | 98 | 20 | 22 | 103353 |
| CH255-559J12 | CU104673.1 | 0 | 0 | 0 | 2000 |
| CH255-397I3 | CU104663.1 | 0 | 0 | 0 | 2000 |
| CH255-469C9 | CU104668.1 | 0 | 0 | 0 | 2635 |
| CH255-56N15 | CU104674.1 | 4 | 2 | 1 | 32416 |
| CH255-114D6 | CU104652.1 | 0 | 0 | 0 | 2000 |
| CH255-58L21 | CU104676.1 | 0 | 0 | 0 | 2000 |
| CH255-351B13 | CT025711.1 | 0 | 0 | 0 | 29819 |
| CH255-354J20 | CT025621.2 | 0 | 0 | 0 | 2000 |
| CH255-336G22 | CT025558.1 | 0 | 0 | 0 | 2000 |
| CH255-191J6 | CU104655.1 | 0 | 0 | 0 | 2000 |
| CH255-206J13 | CU104657.1 | 101 | 14 | 16 | 59844 |
| CH255-529K7 | CU104672.1 | ||||
Variation shown is between the clone listed on that row and the next, in the length of the overlap between the two. Clones are listed in the order of contiguous overlap.
SNPs = single nucleotide polymorphisms.
Comparative analysis
Overall, a high level of conserved synteny is observed between the entire gorilla MHC and the human equivalent as annotated for the PGF cell line (which is the MHC reference haplotype). There are, however, regions where notable differences between the human and gorilla MHC have been identified. These regions are highlighted in Figure 2, which will be referred to during the discussion that follows. In addition to the conservation of functional loci across the region, many processed pseudogenes are also conserved between gorilla and human.
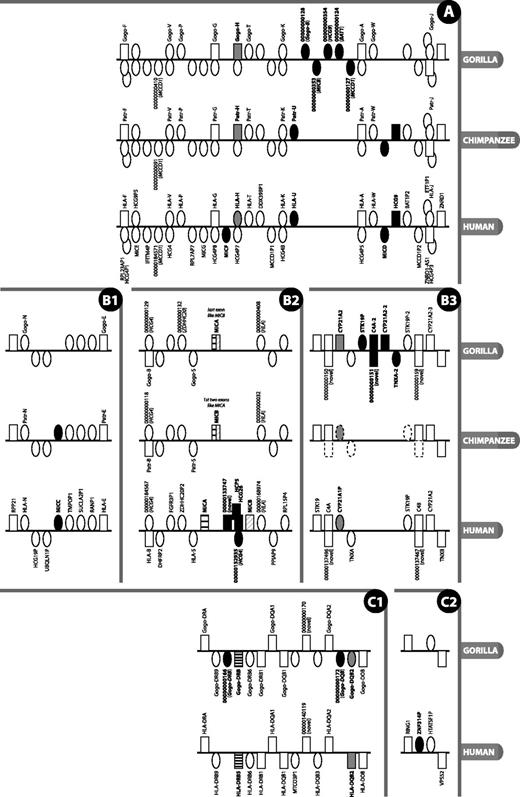
Detailed view of regions of the MHC where there is a difference in gene content or type between gorilla, human reference and chimpanzee. Figure is not to scale. Rectangle = gene; oval = pseudogene; grey fill = type difference (pseudogene versus gene); black fill = gene absent/present in at least one species and not another; black and white striped = not direct orthologue; above line = locus on forward strand (in reference to human chromosomes); below line = locus on reverse strand; stacked = genes overlap or are nested. Gene names are given where available and are only shown for gorilla and chimpanzee when different from human; locus names that appear as numbers with leading zeros are loci without approved nomenclature, with the numbers representing the numerical part of VEGA stable gene IDs (to obtain the full ID, the 11-digit number should be prepended with OTTGORG, OTTPANG or OTTHUMG for gorilla, chimpanzee and human, respectively). An italicised locus name between brackets for a pseudogene indicates the parent gene or gene family of that pseudogene. The loci on the chimpanzee contig in panel B3 are annotated by ENSEMBL (release 70, January 2013), with dotted outlined loci indicating manually determined genes not annotated by ENSEMBL. Section labels A, B and C have been added to allow for easier reference to this figure in the text.
Extended class I
There are very few differences between gorilla and human in the extended class I region. Moving along the chromosome from the telomeric end of the p arm towards the centromere, we find two olfactory receptor loci (OR5V1 and OR10C1) within the olfactory receptor gene cluster that have been classified as coding loci in human but appear to be pseudogenes in gorilla. Conversely, there are two olfactory loci (OR2B4P and OR12D1P) that are classified as pseudogenes in human but have the potential to encode a protein in gorilla (OR2B4 and OR12D1). The total number of olfactory receptor loci within this region, 25, is the same in human and gorilla. This is in broad agreement with the findings of others that the deterioration of the olfactory repertoire of Old World monkeys compared with mouse, New World monkeys (except the howler monkey) and the lemur (a prosimian) has occurred concomitant with the acquisition of full trichromatic colour vision in primates (17). The human olfactory region has evolved by copy number variation through duplication and deletion, and other findings revealed that all deletion alleles were human derived when compared with the chimpanzee reference genome (18). More recently, gorillas have been found to have a functioning olfactory sense that they use to investigate their environment. Thus, olfaction may not be as irrelevant in great apes as has been suggested (19).
Near the boundary with the classical class I region, the human genome carries the single-exon MAS1LP pseudogene as well as MAS1L, a coding gene for a Mas-related multi-pass membrane G-protein–coupled receptor. In gorilla, where MAS1LP is present too, the MAS1L gene encodes, at best, an N-terminally truncated peptide (317 versus 378-amino acids), or is, at worst, a pseudogene. The truncation is caused by a single nucleotide insertion in the gorilla genome in codon 6, just downstream of the equivalent of the human ATG start codon. The next available ATG is 61 codons downstream, with two further ATGs 2 and 4 codons from there; the latter has good Kozak sequence, on par with the upstream human ATG.
Class I—classical: HLA genes and orthologues
The class I genes in gorilla show a high level of conserved synteny with human. The notable differences are confined mainly to small sub-regions, as illustrated in Figure 2A, and will be highlighted as they are discussed.
As expected from the literature, the three classical class I gorilla orthologues of the human HLA-A, -B and -C genes—Gogo-A, -B and -C—are present and appear to be expressed. In our analysis of the chimpanzee BAC sequences from Anzai et al., we confirm that they are expressed there as well (20–22).
Class I—non-classical: HLA genes and orthologues
The human HLA-E, -F and -G genes have protein-coding gorilla and chimpanzee counterparts. The remaining HLA-A paralogues in human are unprocessed pseudogenes, often polymorphic and in varying states of degradation: from full length with eight exons in HLA-H, -K and -J to only single exons in -N and -U (3). Interestingly, in contrast to human, the gorilla and chimpanzee orthologues of HLA-H—Gogo-H and Patr-H—have the potential to encode a full-length 362-amino acid peptide (Figure 2A). HLA-H is a pseudogene in the human reference genome (haplotype PGF) and in haplotypes APD, COX, DBB, MANN, MCF, QBL and SSTO. Pseudogenisation is caused by different frameshift-causing single nucleotide polymorphisms in different haplotypes: in PGF, it is an insertion just after the splice acceptor site of exon 4, and in the other haplotypes, it is a deletion in the middle of exon 4. The pattern of variation within human HLA-H is consistent with it being in neutral linkage with the neighbouring HLA-A paralogues as a result of balancing selection acting on the neighbouring functional HLA loci (23). A Gogo-H-specific transcript has been identified in gorilla, suggesting this locus is indeed transcribed, translated and functional. This gene has been found by others to be expressed in both chimpanzee and gorilla (7, 24). However, Anzai et al. (22) found it to be a pseudogene in chimpanzee, so the locus appears to be haplotypic in chimpanzees. The human orthologue of Gogo-P—HLA-P—has been defined as a transcribed pseudogene in the HLA family, but as yet, there is no evidence that it is transcribed in gorilla. The orthologue has been lost in rhesus macaque (25), but we annotated an orthologue in chimpanzee along with Patr-T and Patr-W (26, 27), as indicated in Figure 2A.
In gorilla, the region between Gogo-K and Gogo-A is of interest in that it is 72.6-kb in length, some six times the 12-kb distance between HLA-K and HLA-A (see Figure 2A). This human, and chimpanzee, 12-kb interval carries just the HLA-A paralogue HLA-U, an unprocessed pseudogene. In gorilla, this region contains five unprocessed pseudogenes—CH255-39I5.5 (OTTGORG00000000128), CH255-39I5.18 (OTTGORG00000000353), CH255-39I5.19 (OTTGORG00000000354), CH255-39I5.6 (OTTGORG00000000124) and CH255-39I5.7 (OTTGORG00000000127)—that appear to be remnants of Gogo-B, MICB, HCG9, BAT1 and MCCD1, respectively; gorilla lacks an HLA-U orthologue. A MIC pseudogene known as MICD and a non-coding locus known as HCG9, which lie between HLA-W and HLA-J in human, are also absent from the corresponding location in gorilla. Interestingly, what appears to be a remnant of HCG9 (pseudogene CH255-39I5.19 or OTTGORG00000000354) is just discernible in gorilla in an upstream location close to MIC pseudogene CH255-39I5.18 between Gogo-K and Gogo-A. A dot plot of human versus gorilla genomic sequence from HLA-F to HLA-J (Supplementary Figure S1) suggests that stretches of sequence within this region have been subject to duplication and re-arrangement after the divergence of gorilla and human. The 48-kb distance between HLA-W and HLA-J in human and chimpanzee is ∼5-kb longer than the corresponding region in gorilla. Despite these differences, overall, the gorilla and human exhibit a high degree of similarity. This is unlike the situation in rhesus macaque where a large expansion of class I genes has occurred between Mamu-G and Mamu-J, the orthologues of HLA-G and HLA-J (28).
Class I: MIC
The human lineages studied to date carry two functional MIC genes—MICA and MICB (29)—and a number of unprocessed pseudogenes (MICC, D, E, F and G) (25, 30). MICA and MICB are thought to have diverged from each other 33-44 million years ago (31), before the divergence of chimpanzee and human. We have found a major difference between human and gorilla in the apparent fusion of the MICA and MICB genes into what we have called MICA, similar to what has been observed in chimpanzee (32) (see Figure 2B2). This single hybrid MICA–MICB gene appears to have originated as a result of a 95-kb deletion in the region compared with human (22, 33), and shows only limited polymorphism between different individuals (32). The fact that the gorilla coding exons 1-5 show greater sequence similarity to human MICA compared with the final 3′ coding exon, which shows greater similarity to MICB. In chimpanzee, it is a slightly different fusion: the first two exons appear to be derived from MICA and the remaining exons from MICB, supports the theory of a deletion between and through the genes, as opposed to deletion of a single gene. This apparent fusion of MICA and MICB in gorilla and chimpanzee is accompanied by the loss of orthologues of the intervening genes (HCP5, an HCG4 pseudogene and HCG26). MIC pseudogene MICC, between UBQLN1P and TMPOP1, is also lost in gorilla, but it is present in chimpanzee (Figure 2B1). This region is clearly subject to both large and small deletions and subject to polymorphism as well. Ando et al. (34) found no linkage disequilibrium between MICB and MICA, but the human MICA and MICB genes exhibit extensive polymorphism (35) as evidenced by the fact that at the time of writing, the IMGT/HLA database (36) lists 84 MICA and 40 MICB alleles. Furthermore, human alleles DRB1*03 and DRB1*07 present with a polymorphism characterised by a 4-bp insertion in MICB, resulting in an extended and altered open reading frame (ORF) at the 3′ end (6). There are instances of Northeast Asian and Native American individuals with the HLA-B*4801 allele haplotype who have lost the MICA locus owing to a large 100-kb genomic deletion. The genomic breakpoints are distinct from those observed in chimpanzee (37, 38). Anzai et al. note that this region is subject to deletion and, on the basis of sequence differences between human and chimpanzee, narrowed down a possible recombination breakpoint to a segment located between the ends of MICA’s second and MICB’s fourth introns. All in all, it is intriguing that a similar-sized deletion involving the same genes (MICA and MICB) has occurred independently in different primate species. The molecular basis of the apparently deletion-prone nature of the segment between MICA and MICB remains to be established, but the presence here of a HERV-L endogenous retrovirus sequence, which contains a 2.5-kb AT-rich insertion in its 5′ long terminal repeat that might serve as a recombination hot spot, could have some involvement (39).
MICF, one of five MIC pseudogenes, which lies between HLA-G and HLA-H in the human MHC, appears to be absent in gorilla (Figure 2A). This is consistent with the chimpanzee (22).
Class I: other
The translation of the gorilla orthologue of PSORS1C1 (psoriasis susceptibility 1 candidate 1) terminates prematurely relative to human and chimpanzee, both of which have genes encoding 152-amino acid proteins. A single nucleotide deletion in a polyC tract in the penultimate coding exon 5 (in codon 39 or 40 of the CDS) of gorilla PSORS1C1 leads to a frameshift in the CDS. This introduces 24 novel C-terminal amino acids in the peptide before a premature termination codon in the last exon. In the genome of human haplotype QBL, the same deletion results in the same potential translation of 63-amino acids. This contrasts with haplotypes COX and PGF, which have PSORS1C1 genes coding for a full-length 152-amino acid peptide. It would be of interest to investigate whether a similar discrepancy exists between different gorilla haplotypes. Further analysis is required to establish whether the gorilla PSORS1C1 locus is functional population-wide or not.
Class II
The class II region generally shows notable divergence between species (40, 41); however, a high degree of conservation exists between gorilla and human in the vicinity of the DRB loci. Only one of the two variants of the C6orf10 gene that have been annotated in human could be annotated in gorilla. The second variant annotated in the human PGF assembly (OTTHUMT00000076177) contains three additional exons, one of which cannot be resolved in gorilla (between gorilla exons 19 and 20).
The gorilla class II region contains one DRA locus—Gogo-DRA—and five DRB loci: Gogo-DRB9 (a pseudogene, as in human), CH255-114D6.4 (OTTGORG00000000166, a small DRB pseudogene fragment not present in human), Gogo-DRB (known as Gogo-DRBY*01 or Gogo-DRB*W8:02) (42), pseudogene Gogo-DRB6 (Gogo-DRB6*01:02N) (43) and Gogo-DRB1 (Gogo-DRB1*10:01) (44). Alleles mentioned above are from the Immuno Polymorphism Database, non-human primate MHC section (45, 46). In human, this region is subject to haplotypic variation (47–49), with different individuals having a different number and/or complement of genes out of the nine DRB genes. The Gogo-DRB gene is located in a position equivalent to HLA-DRB5, but it is not a direct orthologue: it is from a haplotype with a complement of genes slightly different from the human reference. DQB3 exists as a pseudogene fragment, as is the case in human. The Gogo-DQB2 locus terminates prematurely in gorilla compared to human owing to an out-of-frame tandem repeat expansion in exon 3 and has therefore been annotated as a pseudogene (see Figure 2C1). An additional DQB pseudogene fragment—CH255-354J20.5 (OTTGORG00000000172)—lies immediately downstream of Gogo-DQB2. Except for the absence of zinc finger pseudogene ZNF314P (Figure 2C2) and an orthologue of human HCG25, the gene order and content of the remainder of the class II and extended class II in gorilla, from TAP2 to KIFC1, is identical to that in human.
Based on the gene and exon arrangement of DRB genes in this gorilla, the DR type would appear to be DR51 (3). It is difficult to say much about this region in chimpanzee, as this falls outside the area for which clone-based sequence is available. Owing to the repetitive and variable nature of this region, the whole genome shotgun sequence that is available cannot be completely relied on, especially because there are currently many assembly gaps in this region. With this caveat in mind, however, it would appear that chimpanzee has a functional DQB2 gene (ENSPTRG00000023404) like human.
Class III
The genes in the class III region of the gorilla were found to share a high degree of conserved synteny with the human class III genes, and this is in accordance with what others have found in pig, mouse and zebrafish (50–53). One location where differences do occur among the primates, however, is within the RCCX region (54). Early identification of the genetic heterogeneity of this C4 to CYP21 region in primates was made in 1991 (55), when the likely timing of the original duplication was established to be at >30 million years ago. Not only do there seem to have been duplications of this gene block (56) but also an insertion of a 6.5-kb endogenous retrovirus ERV-K into intron 9 of the C4 gene, which has given rise to both short and long forms of this gene (57, 58). Incidentally, this ERV is one of the most biologically active of the endogenous retroviral insertion elements and is possibly still being actively transcribed at a low level in the population (59). The ancestral C4 gene in primates was likely to have been the short C4 gene (57) and because in some humans at least both the derived copies C4A and C4B are found in the long form (4, 60), one can surmise that this insertion pre-dates any duplication event. Results from the many studies of this region in both different primates (55) and in different individuals from the same species (61, 62) show that after the initial duplication, the region has undergone repeated and unequal homologous intragenic crossing over in all the primate species examined (63). No examples of chimpanzee or gorilla RCCX regions studied to date (56, 57, 64), however, have shown the presence of the ERV in any of the copies of the C4 gene, and both species can carry multiple copies of the block (62). Because the ERV is found in African green monkeys, rhesus macaques (56), orangutans and humans in exactly the same intron of their C4 genes (60), it seems more probable that the processes of duplication and homogenisation that have occurred in this region have resulted in the loss of the ERV [perhaps early in the lineages of these two species after the first duplication(s)], rather than that neither species ever carried them (57).
In our gorilla, there appear to be three copies of this module (trimodular structure): we have annotated three C4 loci (C4A, C4A-2 and C4B, all of the same short length), three full-length CYP21A2 loci (CYP21A2, CYP21A2-2, CYP21A2-3), two copies of the TNXA pseudogene and two copies of STK19P (STK19P and STK19P-2) (Figure 2B3). All three copies of CYP21A2 are full length, and thus have the potential for being functional. This is not the case with the human haplotypes studied, where just one copy of CYP21A2 is functional and additional copies appear to be pseudogenes (4). Although in the human haplotypes we have studied we did not find a trimodular structure, it has been found in other individuals (61, 65). There is no chimpanzee BAC sequence for this region, but whole-genome shotgun sequence from the same chimpanzee (‘Clint’) shows it to have a bimodular structure like the human reference (Figure 2B3). Variation in the RCCX cluster has been linked to various autoimmune diseases, such as arthritis (66) and lupus (67).
Conclusion
We have, for the first time, presented the comprehensive annotation of the gorilla MHC genomic region (16). Sequencing was clone based, and therefore of the high quality required for regions of a highly variable and duplicated nature. As can be seen in Figure 3, which shows the RCCX region of the mostly Illumina de novo–assembled whole-genome shotgun sequenced ‘Kamilah’ genome (14) in the ENSEMBL browser, there are several assembly gaps. Also, some predicted gene models (TNXB for example) straddle the gaps and almost certainly merge parts of separate loci (TNXA and TNXB for example) that are not completely presented on the assembly because of mis-assembly and/or assembly gaps. Because of the assembly gap in the middle of the RCCX region, it is not possible to say whether ‘Kamilah’ has a bimodular or trimodular RCCX structure. Quantitative and qualitative module differences have been associated with disease in human (61, 66, 68, 69).

ENSEMBL browser view (release 70, January 2013) of the RCCX cluster and flanking regions of the genome of ‘Kamilah’ (whole-genome shotgun gorilla sequence) showing assembly gaps (white between the blue contigs) and gene models straddling assembly gaps and merging separate fragmented loci (green arrows). See Figure 1 legend for description of features.
For comparison purposes, we have presented updated annotation of the clone-based sequence of the chimpanzee class I MHC (70) sequenced by Anzai et al. (22). This means that at least for the class I MHC region, we have accurate annotation for three closely related primates—human, gorilla and chimpanzee—which allows researchers to compare MHC structure and evolution between, for example, Old World primates and New World monkeys, the latter of which seem to have a less diverse MHC (71).
The genes in the MHC region are of great medical interest (72) because they are critical for the vertebrate immune system. The region’s evolution is driven by the capacity to combat infectious disease, and positive selection operates on MHC loci to maintain variation; hence, the greater the diversity of class I and II molecules (both qualitatively and quantitatively), the greater the possibility of survival of a species (73). Until now, in no other higher primate has the MHC been sequenced and analysed to the same depth as the human, so this study of such a close ancestor should prove a valuable resource that can be expected to advance our understanding of the structure, function, variation and evolution of this complex region in primates. It also adds to the growing body of data on MHC genes and regions (74) in vertebrates in general (75–78).
Materials and methods
Mapping
The BAC clones in this study were chosen from the CHORI-255 library (Children's Hospital Oakland Research Institute, Oakland, CA), in cloning vector pTARBAC2.1, from a blood sample of a heterozygous male Western lowland gorilla (‘Frank’, #327, Gorilla gorilla gorilla) housed at the Lincoln Park Zoo (Chicago) (79). Further details of the BAC library can be found at (80). Screening of the redundant set of library filters was carried out as described by Stewart et al. (81) for the human MHC haplotype study using the human-derived ‘overgo pairs’ probes. A total of 333 positive BAC clones were placed in a new array, which was used to generate 70 identical filters that were then probed individually with the full set of ‘overgo pairs’. These clones were then BAC-end sequenced, restriction (HindIII) fingerprinted and mapped to a single contig using the methods described by Stewart et al. The high-quality BAC-end sequences were matched back to the human MHC in ENSEMBL with BLAST (Basic Local Alignment Search Tool) to enable a minimally overlapping tile-path of 29 BAC clones to be selected for sequencing.
Sequencing
The entire gorilla MHC region constituting a tile-path of 29 BACs was sequenced at the Wellcome Trust Sanger Institute (United Kingdom). Sub-cloning, capillary sequencing and finishing were performed using the standard procedures in operation at the Sanger Institute at the time and as described in summary in the headers of the International Nucleotide Sequence Database submissions for these BAC sequences.
Sequence annotation and analysis
Manual annotation was uniformly performed on the entire gorilla MHC sequence by the Wellcome Trust Sanger Institute HAVANA team as follows. The finished gorilla genomic sequence was analysed using an automatic ENSEMBL pipeline (82) with modifications to aid the manual curation process. The G + C content of each clone sequence was analysed, and putative CpG islands marked. Interspersed repeats were detected by RepeatMasker (83) using the mammalian library along with primate-specific repeats submitted to the INSDC (International Sequence Database Collaboration of DNA Data Bank of Japan (DDBJ), European Nucleotide Archive (ENA) and GenBank) databases, and simple repeats were found using Tandem Repeats Finder (84). The combined repeat types were used to mask the full sequence. This masked sequence was searched for matches to vertebrate cDNAs and expressed sequence tags (ESTs) using WU-BLASTN (85), and matches were refined and cleaned up using EST2_GENOME (86). A protein database combining non-redundant data from SwissProt and TrEMBL was searched using WU-BLASTX (85). Ab initio gene structures were predicted using FGENESH (87) and GENSCAN (88). Based on the aforementioned analysis, gene or transcript models were manually annotated according to the ENCODE Genome Annotation Assessment Project (EGASP) guidelines (89). The gene categories used were as described on the VEGA website (15, 90) and the annotation guidelines available from the HAVANA website (91): known genes are identical to known gorilla cDNA or protein sequences or are orthologues of known human loci; novel CDS loci have an ORF and are identical to spliced ESTs and/or have some similarity to other genes or proteins; novel transcripts are similar to novel CDS loci but no ORF can be determined unambiguously; putative genes are identical to spliced ESTs, but do not contain an ORF; and pseudogenes are non-functional copies of known or novel loci with coding regions disrupted by premature stop codons and/or frameshifts.
Funding
This work was supported by the Wellcome Trust. The Wellcome Trust (098051).
Conflict of interest. None declared.
Acknowledgements
The author would like to thank the members of the sequencing, sequence finishing and HAVANA teams for their invaluable contribution.
References
Author notes
Citation details: Wilming,L.G., Hart,E.A., Coggill,P.C., et al. Sequencing and comparative analysis of the gorilla MHC genomic sequence. Database (2013) Vol. 2013: article ID bat011; doi:10.1093/database/bat011



