-
PDF
- Split View
-
Views
-
Cite
Cite
Steve Fischer, Cristina Aurrecoechea, Brian P. Brunk, Xin Gao, Omar S. Harb, Eileen T. Kraemer, Cary Pennington, Charles Treatman, Jessica C. Kissinger, David S. Roos, Christian J. Stoeckert, The strategies WDK: a graphical search interface and web development kit for functional genomics databases, Database, Volume 2011, 2011, bar027, https://doi.org/10.1093/database/bar027
Close - Share Icon Share
Abstract
Web sites associated with the Eukaryotic Pathogen Bioinformatics Resource Center (EuPathDB.org) have recently introduced a graphical user interface, the Strategies WDK, intended to make advanced searching and set and interval operations easy and accessible to all users. With a design guided by usability studies, the system helps motivate researchers to perform dynamic computational experiments and explore relationships across data sets. For example, PlasmoDB users seeking novel therapeutic targets may wish to locate putative enzymes that distinguish pathogens from their hosts, and that are expressed during appropriate developmental stages. When a researcher runs one of the approximately 100 searches available on the site, the search is presented as a first step in a strategy. The strategy is extended by running additional searches, which are combined with set operators (union, intersect or minus), or genomic interval operators (overlap, contains). A graphical display uses Venn diagrams to make the strategy’s flow obvious. The interface facilitates interactive adjustment of the component searches with changes propagating forward through the strategy. Users may save their strategies, creating protocols that can be shared with colleagues. The strategy system has now been deployed on all EuPathDB databases, and successfully deployed by other projects. The Strategies WDK uses a configurable MVC architecture that is compatible with most genomics and biological warehouse databases, and is available for download at code.google.com/p/strategies-wdk.
Database URL: www.eupathdb.org
Introduction
The potential for discovery through data mining is governed, in part, by the scale and scope of available data sets, and the range of searches available. Many genomics databases support multiple organisms, and integrate a wide range of data sets beyond the genome and gene models. Reference organisms may include curated or automated functional associations, transcript and protein expression data (based on many technologies), pathway associations, phenotype information, polymorphism data, comparative genomics analyses, protein structure information, cellular location, etc. Each data type, and each data set, makes additional dimensions available for mining. For example, one of the EuPathDB sites, PlasmoDB.org (1), the Plasmodium Genomics Resource, offers nearly 100 searches and a total of approximately 250 parameters. Many other sites, such as the Saccharomyces Genome Database (SGD; www.yeastgenome.org) (2) or the Ensembl website (www.ensembl.org) (3), support advanced multiparameter searches, but the full dimensionality of the available data is most useful only if the results of searches can be logically combined.
EuPathDB sites (4) have offered intersect, union and minus operations since 2001, but until recently users had to go to the Query History page to perform these operations. A simple text box allowed them to specify expressions like (1 AND 2) OR 3, which would find the intersection of Results 1 and 2 and union it with Result 3. While advanced users took advantage of this interface, most ignored it, limiting themselves to simple searching.
This raised the challenge of designing a graphical interface that helps users manage the breadth of available searches and encourages them to combine the search results. Our solution is a configurable web framework for database searching called the Strategies Web Development Kit (WDK) that includes a new strategies web application as its graphical user interface (Figure 1). We developed the Strategies WDK with formal feedback from users at EuPathDB workshops and focus groups of investigators using EuPathDB sites. It is a mature system that has been in production since 2001, with the new GUI in production since June 2009. The Strategies WDK has also been adopted and deployed by other projects (the TB Database: www.tbdb.org, and the Beta Cell Biology Consortium: genomics.betacell.org). The system is open source under the GNU LGPL and is available at code.google.com/p/strategies-wdk.
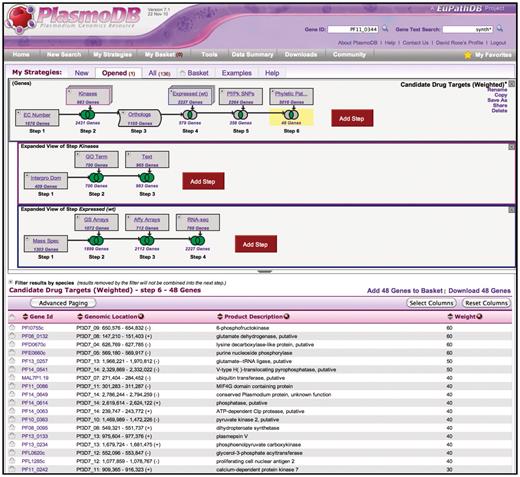
The WDK Strategies system in PlasmoDB.org. The top panel shows a strategy with a succession of gene searches (strategy steps) proceeding from left to right, combined using set operators (e.g. union, intersection) indicated by Venn diagrams. Steps 2 and 4 are nested steps, indicated by an overlapping box icon with colored border. They are expanded as sub-strategies in the indented panels below, where their component steps are visible. Step 3 is a transform expanding the results from Step 2 to include orthologous genes. Strategies may be named and saved for future reference, refinement, or sharing with others. The bottom half of the display is the summary table which shows a page of results, where each row is one gene found. By default this display shows the result of the strategy as a whole (the last step so far), but clicking on any step box or Venn diagram highlights that box in yellow and shows the intermediate result at that point. Any of these results are downloadable. The summary table can be customized by adding any of approximately 50 columns, and by sorting, filtering and paging.
Features
Overview
The Strategies WDK provides users with a richly featured search system on a single web application page. Users remain on that page to run and interact with all active searches. An initial search becomes the first step of a Strategy, and additional searches are added as needed to extend the strategy refining the final result. Formally, the strategy can be represented as a tree, where the leaves are searches, and internal nodes are parameterized operations that combine or transform results. Available combining options include ‘set operations’ (intersect, union and minus) and ‘interval operations’ (overlap or containment on the genome), both of which take in two result sets and produce a combined result. ‘Transforms’ take in one result set and produce one transformed set as output, such as finding orthologs of the input set. In the display, strategies are shown as linear, but may be nested (see below), allowing strategies to be arbitrarily complex.
User experience
All searches on EuPathDB sites are run within the Strategy system. This is convenient for users who want to run a simple search, for those who start simple and realize that refining their search will yield more precise results or for users who have devised a complex plan for data mining. By way of example, a user may wish to identify Plasmodium genes that could be involved in modulating host cell signaling pathways: ‘putative kinases that are secreted (or membrane bound), and also supported by experimental transcript data’.
Building a strategy
The user begins a search from toolbar pulldown menus or menus on the home page, and is offered a large set of searches organized by type to facilitate navigation. Each search is orthogonal to the others, and returns records of a single type. PlasmoDB v7.0 offers approximately 60 gene searches, and approximately 40 additional searches for isolates, genomic sequences, single nucleotide polymorphisms (SNPs), expressed sequence tags (ESTs), transcript assemblies, ORFs and SAGE tags. (Searches are designed by the installer; see System Design and Installation below.) In Figure 2, the user has run a simple, single-step search for genes annotated with the Gene Ontology (GO) term ‘kinase activity’ (http://www.geneontology.org) (5). Results are automatically displayed in the strategy system. The ‘Strategy Panel’ (top) provides the user a graphical layout of the strategy, which grows as additional searches are performed (cf. Figure 1). After an initial search, the panel displays a box at the left, representing that search as ‘Step 1’ in the strategy. In the example shown, the first step is labeled ‘GO Term’ and ‘700 Genes’, indicating the type of search and the result count. The bottom half of the results page is the ‘Summary Table’, a tabular display of the records found, which in this case is a list of genes.
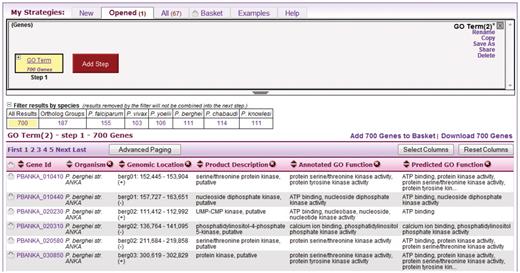
A single-step strategy. This is what the user sees after running a new search, in this case a search for genes by GO term. The user clicks the Add Step button (red) to add another search to the strategy.
To the right of the Step 1 box is a conspicuous ‘Add Step’ button, which the user can click to grow the strategy. Doing so pops up the Add Step dialog box presenting a menu of options for the next step (Figure 3a). The user will typically run another search, choosing from among all searches available on the site. S/he can also run a transform or embed a saved strategy as a nested step. In this example, the user has opted to search for Genes based on the presence of Transmembrane (TM) domains (one of several subcellular localization queries), and the appropriate search form now appears in the Add Step dialog box (Figure 3b). The user specifies the parameters for the search and, at the bottom of the display, chooses a Venn diagram indicating how this new step should be combined with the accrued results from previous steps. Figure 3c shows the combined result (77 genes in the example shown).
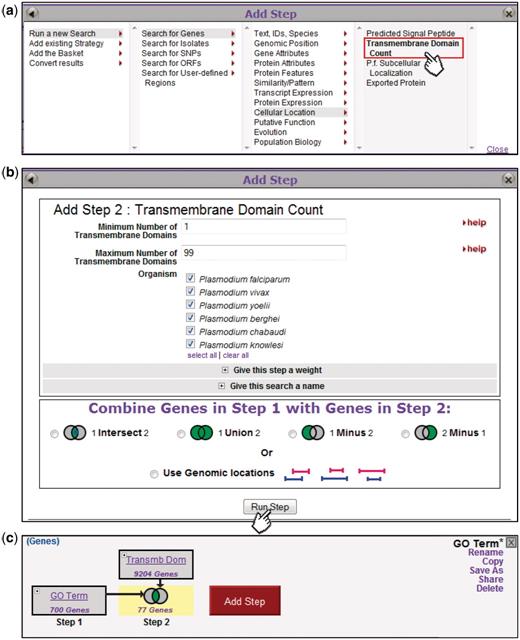
Adding steps to build a more complex strategy. Clicking on the Add Step button (Fig. 2) brings up an Add Step dialog box (a), from which the user selects which search to run. This brings up a display showing parameters for that search (b, top) and options for combining with the previous step (b, bottom). The new step is added to the strategy (c), and the combined result highlighted and returned as a list in the Summary Display (data not shown).
Strategies are dynamic
Results generated from a strategy are updated dynamically as new steps are added, existing steps changed or transformations applied. Clicking on a step highlights that box in yellow, and displays the results of that search in the summary table (cf. Figure 1). Clicking on a Venn diagram displays the results of the accrued operations to that point (cf. Figure 2). This allows the user to examine the results of intermediate components in the search strategy, and adjust them if necessary. Figure 4a shows three steps (the user has added a third step to limit the result to genes with EST evidence). Clicking on the ‘+’ icon of Step 2 opens the step’s ‘action menu’ (Figure 4b). The user sees the details of the search’s parameters and is offered a number of actions such as view, insert before and delete.
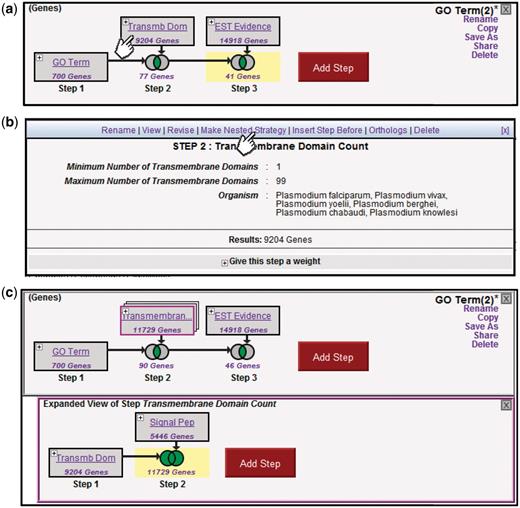
Creating nested Strategies. Clicking on the + icon in Step 2 of the three-step strategy shown (a) brings up a menu displaying the details of this step (b), and a set of available actions (Rename, View, Revise, Make Nested Strategy, Insert Step Before, Orthologs, Delete). Clicking on Make Nested Strategy expands the step into its own panel (c), where the user can add additional steps that become nested within the second step in the top panel (TB domain and/or signal peptide in the example shown).
Figure 4c illustrates additional options for improving a strategy. A search for genes predicted to encode proteins with TM domains is expanded to those with TM domains ± (union) signal peptides. Clicking on ‘Make Nested Strategy’ in the actions menu for Step 2 expands this step into its own panel below the main panel. By allowing such embedded substrategies, the strategies system supports nested set operations. Adding a step in the bottom panel to search for genes returned by a SignalP search, and using the Venn diagram to construct the union of these results with the TM step, yields an additional 2525 genes in the entire database (11 729 total), contributing four more genes to the final result in the top panel. The action menu also provides a revise option, allowing users to adjust the parameters of any step in the strategy, with the updated results propagated forward through the strategy. For example, the user might revise Step 1 to change the GO term from ‘kinase activity’ to ‘phosphatase activity’, identifying genes of complementary function.
Other dynamic aspects of the Strategy system allow users to add, remove, sort and reposition columns in the summary table. PlasmoDB provides approximately 55 attributes that may be displayed as columns for gene results (gene length, predicted protein molecular weight, time of maximal expression, etc). Also in Figure 2, the ‘quick filters’ section seen between the strategy panel and summary table lets users apply filters to the results with a single click. For example, the user can limit the view of the result to a selected organism or strain.
(The strategy built in the above example is shared. To view and edit a copy of it, go to plasmodb.org/plasmo/im.do?s=aea5452877157ff5.)
Additional features
All searches in PlasmoDB are compatible with the strategies system, including BLAST (6) and motif searches (which on many sites are returned as static pages only). In the WDK, these are treated as searches that can be added as steps in a strategy, contributing genes to the larger data mining process. Similarly, transformations such as finding the orthologs of a gene set produce strategy-compatible results.
Using interval operations as an alternative to set operations, users can combine search results based on the locations within the genome sequence. For example, strategies identifying genes of interest, as illustrated above, may be augmented by adding a search for DNA motifs, using overlap to combine the results with the genes, thereby identifying all genes harboring a particular DNA motif within 500 nt upstream. Another user might wish to identify SNPs contained within non-protein-coding genes. Yet another might wish to examine divergently transcribed genes, i.e. opposite strand genes whose upstream regions overlap. These kinds of operations will be familiar to users of systems such as Galaxy (galaxy.psu.edu) (7).
Each search also offers the user an optional weight parameter that may be used for sorting. For example, one might choose to apply different weights to different forms of expression evidence, weighting protein-level expression most highly (e.g. a weight of 30), followed by microarray or RNA-seq evidence (e.g. 20), and applying a lower weight to noisy sources of evidence such as SAGE tags (e.g. 10). In the final result, genes supported by all of these searches would have a weight of 60, those found only by protein expression will have a weight of 30 and those supported only by SAGE tag evidence 10. Sorting by weight allows the user to identify the best candidates, even if no candidate is supported by all lines of evidence. The user can apply a large number of filters without reducing the resulting set to zero: all records found are retained, but the least interesting fall to the bottom. In contrast, the intersection operation can result in overfiltering, and lost candidates. A similar system is used for target prioritization by the TDR Targets database (tdrtargets.org) (8).
A basket feature allows users to collect individual records or set of records by clicking on a distinctive basket icon, which accompanies IDs displayed throughout the interface (cf. summary table in Figure 2). Records in the basket can be downloaded as a report, operated on using analysis tools (e.g. for multiple sequence alignment), or added as a step in a strategy for further operations. The favorites feature lets users hand-select individual records for quick access.
Strategies can be named, saved and shared with others using a unique URL. When a colleague opens the URL, a copy of the shared strategy will appear in his or her strategy workspace, and s/he can modify or interact with the strategy without affecting the original. The strategy system also offers a ‘strategies browser’ where the user can examine saved strategies. Strategies are saved by default, providing users who have logged in with a permanent record of their query history, except for those that they choose to delete. Saved strategies can be embedded as modules in other strategies.
Advanced strategies
Using a larger number of steps and substrategies, researchers can develop very specific search strategies. The advanced strategy in Figure 1 finds genes that are candidate drug targets in the malaria parasite, Plasmodium falciparum. The top panel is the main strategy, and the lower panels are the substrategies nested into Steps 2 and 4, respectively. Step 1 identifies genes from any Plasmodium species annotated with an EC number [The Enzyme Commission number is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze (9)], i.e. with evidence of being an enzyme. Because kinases are under-represented in EC annotation, it is unioned with Step 2 which is a substrategy that further searches for kinases. The substrategy uses union to find genes with InterPro domains annotated as kinases, genes annotated with GO Term ‘kinase activity’ and genes with comments and notes matching the term ‘kinase’. The result after Step 2 is 2431 Plasmodium genes with evidence of being an enzyme or specifically a kinase. By selecting that result, the user would see that 998 of those genes are from P. falciparum, with the rest from various other Plasmodium species. Step 3 is an ortholog transform that finds P. falciparum orthologs of any of the 2431 genes in Step 2 (it also removes non-P.falciparum genes from the result). The result is 1105 P. falciparum genes, indicating that the ortholog transform added 107 P. falciparum genes. Step 4 is another substrategy. In its panel, the user has unioned genes showing differential expression in any of four experiments (mass spectrometry, glass slide, microarray and RNA-seq) from the trophozoite life cycle stage, when the parasite is most vulnerable to drugs. While not visible in the panel, each of these steps has been given a weight. The mass spectrometry search has a weight of 30, whereas the others have a weight of 10. The union operation sums weights, so genes with multiple lines of expression evidence will have higher weights in the final result. Step 5 uses a SNP search to restrict the result to genes that have a low rate of non-synonomous SNPs, i.e. that are under purifying selection, so may be essential to the parasite. Step 6 uses an orthology-based phylogenetic profile search to keep only genes that are present in the parasite but not in humans, to ensure that the drug does not harm the host. In the final result shown in the summary table, there are 48 genes ordered by their weight, i.e. the degree of evidence they have for desired expression. In sum, they are an inclusive set of enzymes and kinases with significant expression in a vulnerable life stage that are essential to the parasite yet absent in the host. (This advanced strategy is shared at plasmodb.org/plasmo/im.do?s=1b216883e2065ed0.)
System design and installation
The Strategies WDK system has been in production on EuPathDB websites and in continuous development for 9 years (formerly as the GUS Web Development Kit) (10). The new Strategies interface makes use of current web application technologies such as AJAX and JQuery (see the View section below).
Model
The core system is a Model-View-Controller (MVC) (11, 12) framework (Figure 5) built on an existing application database. The Model for a site is configured in XML and defines data transfer objects (13, 14) called records. These are coarse-grained entities that users will see. For example, EuPathDB sites have records for Genes, SNPs, etc. A record specifies a transformation from fine-grained relational tables in the database into its coarser view. The structure of a record is simple. Its primary key is a unique identifier for the record. Attributes are values that the record has only one of, for example, a gene’s genomic sequence or its genomic location. Tables contain columnar values that the record has many of, for example, a gene’s exons (and their locations), or GO associations (including GO ID, term name and evidence code). Attribute and table values are served to the record object by Structured Query Language (SQL) (when given the record’s primary key). For systems that do not already have an object layer, WDK records are a versatile way to serve data to the front-end.
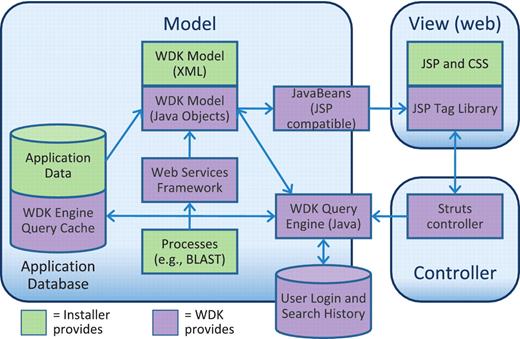
The Strategies WDK MVC design. Scientific data is stored in an application database of any design (supported vendors are Oracle and PostgreSQL). The WDK’s query cache can reside in that database or a separate one. The WDK Model is specified in XML and instantiated into Java objects that represent records and searches. The WDK query engine uses the Java model to perform searches against the application database or using a web services framework. The Java model is made available to the front-end through a JSP-compatible JavaBeans API. The View is primarily comprised of JSP, JSP tags and CSS, and also uses JavaScript, JQuery and AJAX.
The model also specifies searches. A search is configured to have parameters and return a set of records of a specific type. For example, PlasmoDB has a Genes with EST Evidence search with parameters for library, overlap and score. The search executes SQL or a web service call and returns primary keys for the matching records. The WDK displays the results to the user, merging into the display attributes from the record as additional columns (see the ‘User Experience’ section above). It maintains recent search results in a disk cache to facilitate interactive paging and sorting. User logon and search history is maintained in a dedicated user database.
View
The view component of the Strategies WDK displays record pages, search pages and the Strategies user interface, as configured in the model (described above). Each record and search in the model gets a default view. The default record page displays a record’s attributes and tables; the default search page displays a form for the search, including its parameters and help text. Installers can override default pages with customized versions. For example, the PlasmoDB Gene page is customized. Some searches may require customized forms if one or more parameters need a dedicated GUI. The Strategies interface, including the graphics, popup dialogs, column functionality and Strategies Browser are built-in.
The view uses the Java Server Pages (JSP) framework (java.sun.com/products/jsp). JSP pages get a handle on the WDK model through the model’s JavaBeans API. The pages also use JavaScript (15), AJAX (Asynchronous Javascript and XML) (16) and the JQuery (jquery.com) libraries. These technologies retrieve subpage packets without reloading, enabling an application-like experience. User interaction with a Strategy remains in one html page, including adding steps, revising, sharing, sorting, etc. Similarly, search pages have dynamic widgets for gathering parameter values that offer auto-complete, expandable tree values and dependent parameters whose allowed values change as the user makes choices for related parameter values.
Controller
The WDK controller is built on Apache Struts (struts.apache.org). It uses standard action handling appropriate for a web site and web application. It also provides the mechanism for customization by looking in specified directories for custom JSP pages, using those instead of default pages.
Installation requirements
The WDK model and query processing engine is written in Java (Version 1.5 or newer required) and uses a relational database for set processing and for storing user history data. Supported DBMSs are Oracle (10 and 11g) and PostgreSQL (9.0). The model runs in UNIX (tested in Linux CentOS 5.5 and 5.6). The controller runs in Struts (1.2) and the view in Tomcat (5.5 and 6.0) (tomcat.apache.org).
The Strategies WDK has few restrictions on the backend data model. It does not have domain specificity, so the data modeled is unconstrained, except that it must be amenable to representation as records. The installer designs the model appropriate for his or her application and provides hooks to the needed data. The model requires that record attribute and table values be available through SQL. Queries that serve attribute values are performance sensitive so might require dedicated denormalized tables. Record table values are used in download reports and in serving data to record pages. Existing sites may have record pages they are happy with and could opt to forego detailed reports, so table values are optional. Searches can be implemented in SQL directly against a relational database containing application data, or can run through a web service to SOAP (www.w3.org/TR/soap) clients that meet its search API. Record table values also must be accessed directly by SQL.
The WDK includes a set of tools to assist in the model configuration process, including a build system, command line testing framework, an automated testing suite and performance monitoring tools.
Integrating the WDK into an existing front-end or customizing it with a branded look may involve the skills of a web developer. The WDK template site is a working site (strategieswdk.gusdb.org/templatesite), which is delivered with the software. It can be used as a template to accelerate site installation, configuration and customization.
Discussion
Actual usage
To get a sense of the system’s actual usage, we analyzed the PlasmoDB user database for patterns of use. (We did so in aggregate and anonymously, to protect user privacy and methodologies.) In 15-month period, after the system was released (1 July 2009) PlasmoDB users issued 180 000 searches. We found that these can be divided into two basic use cases. The main use case is simple searches. Ninety-one percent of the searches are in single-step strategies, i.e. they do not use the combine feature. Of those, 85% are keyword, ID or BLAST searches. This suggests that a primary use case is a quick search for a set of genes based on text or sequence.
The second use case is data mining. Approximately 9% of the searches (16 000) are part of a multistep strategy, with an average of three steps per strategy. Of these multistep strategies, 45% use features that are particular strengths of the strategies system: union, minus, transforms or nesting. (55% use only intersect, which is more widely available on genomics sites.) About 20% have four or more steps. The revise action was used to refine strategy parameters on about 25% of the steps in the strategies.
Users who log in give us the opportunity to examine (in aggregate) usage patterns of individuals across sessions. (Guest user data is per session only). Of all the searches (9700), 5% were performed by 344 logged in users. Of these searches, 30% were in multistep strategies (three times higher than for guest users). Forty-five percent of logged in users used multistep strategies, with an average of six strategies per user. Seventy percent of these users used advanced strategy features.
While the first use case is dominated by text and sequence searches, the data mining use case favors functional genomics searches such as transcript expression and cellular localization. Excluding keyword, ID and BLAST searches, 33% of all searches are in multistep strategies. Of the 100 searches available in PlasmoDB, 15% are used more in multistep strategies than in single-step.
The patterns of use in the data mining use case suggest that a significant proportion of PlasmoDB users are taking advantage of the Strategies system for complex searching a significant percent of the time, and when they do, they exploit its advanced features.
Related systems
There are a number of features that characterize a sophisticated search interface (Table 1). We surveyed well-known sites that have advanced searching and present our understanding of the availability of those features on their sites. [Some sites not included in the survey, such as WormBase (wormbase.org) (17), RGD (rgd.mcw.edu) (18) and MGD (www.informatics.jax.org) (19), use search systems such as BioMart (www.biomart.org) (20) or InterMine (intermine.org) that are represented by other sites in the survey.] While many genomics database sites offer advanced searching, none besides EuPathDB has a graphical interface that gives users arbitrary set operations, viewing of intermediary results, a diagram of the search logic and interactive search parameter revision (Table 1, row a).
| . | EcoCyc . | Ensembl MartView . | Entrez Gene . | FlyBase . | FlyMine . | SGD . | Strategies WDK . |
|---|---|---|---|---|---|---|---|
| (a) Graphical interface for set operations and interactively revising constituent searches | Yes | ||||||
| (b) Easy to search using simple filters | Yes | Yes | Yes | Yes | |||
| (c) Controlled vocabulary support | Yes | Yes | Yes | Yes | |||
| (d) Filters with multiple parameters | Yes | Yes | |||||
| (e) Users can explore the schema and design arbitrary queries | Yes | Yes | |||||
| (f) Advanced searches developed by experts | Yes | Yes | Yes | ||||
| (g) Set operations including AND, OR and NOT | Yes | Yes | Yes | Yes | Yes | ||
| (h) Nested set operations | Yes | Yes | Yes | ||||
| (i) Weighted searches | Yes | ||||||
| (j) Interval operations | Yes | ||||||
| (k) Transforms/conversions | Yes | Yes | Yes | ||||
| (l) Sharing full power search protocols with colleagues | Yes | ||||||
| (m) Configurable columns | Yes | Yes | Yes | Yes | |||
| (n) Sortable columns | Yes | Yes | Yes | Yes | |||
| (o) Hand-picked sets and sending sets to tools | Yes | Yes | Yes | ||||
| (p) Summary analysis of columns (e.g. histogram of GO terms) | Yes | Yes | Yes |
| . | EcoCyc . | Ensembl MartView . | Entrez Gene . | FlyBase . | FlyMine . | SGD . | Strategies WDK . |
|---|---|---|---|---|---|---|---|
| (a) Graphical interface for set operations and interactively revising constituent searches | Yes | ||||||
| (b) Easy to search using simple filters | Yes | Yes | Yes | Yes | |||
| (c) Controlled vocabulary support | Yes | Yes | Yes | Yes | |||
| (d) Filters with multiple parameters | Yes | Yes | |||||
| (e) Users can explore the schema and design arbitrary queries | Yes | Yes | |||||
| (f) Advanced searches developed by experts | Yes | Yes | Yes | ||||
| (g) Set operations including AND, OR and NOT | Yes | Yes | Yes | Yes | Yes | ||
| (h) Nested set operations | Yes | Yes | Yes | ||||
| (i) Weighted searches | Yes | ||||||
| (j) Interval operations | Yes | ||||||
| (k) Transforms/conversions | Yes | Yes | Yes | ||||
| (l) Sharing full power search protocols with colleagues | Yes | ||||||
| (m) Configurable columns | Yes | Yes | Yes | Yes | |||
| (n) Sortable columns | Yes | Yes | Yes | Yes | |||
| (o) Hand-picked sets and sending sets to tools | Yes | Yes | Yes | ||||
| (p) Summary analysis of columns (e.g. histogram of GO terms) | Yes | Yes | Yes |
| . | EcoCyc . | Ensembl MartView . | Entrez Gene . | FlyBase . | FlyMine . | SGD . | Strategies WDK . |
|---|---|---|---|---|---|---|---|
| (a) Graphical interface for set operations and interactively revising constituent searches | Yes | ||||||
| (b) Easy to search using simple filters | Yes | Yes | Yes | Yes | |||
| (c) Controlled vocabulary support | Yes | Yes | Yes | Yes | |||
| (d) Filters with multiple parameters | Yes | Yes | |||||
| (e) Users can explore the schema and design arbitrary queries | Yes | Yes | |||||
| (f) Advanced searches developed by experts | Yes | Yes | Yes | ||||
| (g) Set operations including AND, OR and NOT | Yes | Yes | Yes | Yes | Yes | ||
| (h) Nested set operations | Yes | Yes | Yes | ||||
| (i) Weighted searches | Yes | ||||||
| (j) Interval operations | Yes | ||||||
| (k) Transforms/conversions | Yes | Yes | Yes | ||||
| (l) Sharing full power search protocols with colleagues | Yes | ||||||
| (m) Configurable columns | Yes | Yes | Yes | Yes | |||
| (n) Sortable columns | Yes | Yes | Yes | Yes | |||
| (o) Hand-picked sets and sending sets to tools | Yes | Yes | Yes | ||||
| (p) Summary analysis of columns (e.g. histogram of GO terms) | Yes | Yes | Yes |
| . | EcoCyc . | Ensembl MartView . | Entrez Gene . | FlyBase . | FlyMine . | SGD . | Strategies WDK . |
|---|---|---|---|---|---|---|---|
| (a) Graphical interface for set operations and interactively revising constituent searches | Yes | ||||||
| (b) Easy to search using simple filters | Yes | Yes | Yes | Yes | |||
| (c) Controlled vocabulary support | Yes | Yes | Yes | Yes | |||
| (d) Filters with multiple parameters | Yes | Yes | |||||
| (e) Users can explore the schema and design arbitrary queries | Yes | Yes | |||||
| (f) Advanced searches developed by experts | Yes | Yes | Yes | ||||
| (g) Set operations including AND, OR and NOT | Yes | Yes | Yes | Yes | Yes | ||
| (h) Nested set operations | Yes | Yes | Yes | ||||
| (i) Weighted searches | Yes | ||||||
| (j) Interval operations | Yes | ||||||
| (k) Transforms/conversions | Yes | Yes | Yes | ||||
| (l) Sharing full power search protocols with colleagues | Yes | ||||||
| (m) Configurable columns | Yes | Yes | Yes | Yes | |||
| (n) Sortable columns | Yes | Yes | Yes | Yes | |||
| (o) Hand-picked sets and sending sets to tools | Yes | Yes | Yes | ||||
| (p) Summary analysis of columns (e.g. histogram of GO terms) | Yes | Yes | Yes |
A number of sites offer an easy-to-use interface to select a series of filters to apply to the search (Table 1, row b). Ensembl MartView (www.ensembl.org) (3) (based on BioMart) may be the best known of these. Multiple filters are available on a single page and are separated in some fashion for clarity. The sites differ in whether and how consistently they offer menus or auto-completion for parameters that could be backed by a controlled vocabulary, such as GO terms, rather than expecting users to know terms and type them correctly (Table 1, row c). EuPathDB does not offer multiple filters on a single page, instead offering a menu of individual searches. The multifilter page has the advantage that the user sees a number of filters at one time and can quickly set a few. However, this approach does not scale well. PlasmoDB, for example, has too many searches to fit comfortably into a single page. Another disadvantage is that the filters apply intersections which, when done successively, may winnow a result to nothing. The user may need to go back and forth to the multifilters page making adjustments to get desired results. [The TDR Targets (tdrtargets.org) (8) database solves this problem on its multifilter page by using unions and weights instead of intersection.]
Some multifilter page implementations allow filters with multiple parameters (Table 1, row d). For example, the Marker filter in Ensembl’s MartView has two parameters, start and stop. EuPathDB accomplishes this by giving each search a dedicated page, which handles the complexity needed for the search. Some EuPathDB searches have numerous parameters, many with values that change depending on user choices for other parameters. The EuPathDB pages also include help and citations for the data source, which none of the other sites provide, and which would be a challenge in the multifilter page approach.
FlyMine (www.flymine.org) (21) and EcoCyc (ecocyc.org) (22) provide tools to navigate database objects directly and form queries on them (Table 1, row e). This has the advantage of opening up the database to arbitrary exploration so users can invent their own queries, providing potentially powerful data mining. A downside is that it may require significant user expertise to successfully develop queries (navigating schemas is notoriously difficult without extensive documentation). EuPathDB does not offer object navigation, relying instead on a rich set of available searches developed by in-house experts and driven by user needs (Table 1, row f). Rare uses cases may not be covered, but the majority that are covered are easy to use. FlyMine solves the problem with their Template system in which experts provide pre-configured queries as a user-friendly layer.
The most effective interfaces give users access to the full complement of set operations. As mentioned above, a series of filters only provides intersection (AND). EuPathDB and more than half the surveyed sites additionally give users union and subtraction (OR and NOT) (Table 1, row g). Fewer of the sites allow nested set operations [(A AND B) OR (C AND D)] as demonstrated in Figure 1 (Table 1, row h). Entrez Gene (www.ncbi.nlm.nih.gov/gene) (23) offers this in a text expression interface, while FlyMine uses point and click but requires users to build up intermediary results. EuPathDB achieves this with nested strategies. Of the surveyed sites, only EuPathDB augments standard set operations with weights (Table 1, row i) (a feature also found on the TDR Targets site). EuPathDB is also the only site to offer interval operations such as ‘Find Genes in set A whose upstream region overlaps SNPs from set B’. (Table 1, row j). Another available operation is what EuPathDB calls ‘transforms’ (Table 1, row k). These have one set in and one set out. FlyMine and FlyBase (flybase.org) (24) call these ‘conversions’. (Biozon www.biozon.org (25) has a particularly interesting version in which sets are transformed based on similarity analysis such as finding genes with similar expression profiles.)
While some of the sites allow users to export and import search specifications for saving and sharing, only EuPathDB does so in a way that retains the power of the search system (Table 1, row l). For example, Entrez Gene users can save an expression that captures the logic of their complex search, but if the expression references previous searches in the history then the expression is not useful to others. FlyMine and FlyBase export text versions of a query but neither format supports full set operations, including nesting.
Search results are displayed in a table of some kind. Across sites, these displays vary in their sophistication. About half of the sites allow the users to configure which columns are displayed, choosing from a potentially large list (Table 1, row m). This lets users see at a glance varied information about their result set. Another group of sites allows users to sort the columns (Table 1, row n). This combines with the former feature to let users bring subsets into the fore. It also combines well with weighted searches, as the highest scoring results can be brought to the top. A few of the sites let users hand-pick rows from the results to submit to a further analysis (Table 1, row o). EuPathDB calls this the ‘basket’ while FlyMine and FlyBase call them ‘lists’. Some sites, EuPathDB excluded, offer summary analysis of a selected column (Table 1, row p). For example, SGD users can analyze the GO terms column to find statistically enriched GO terms. On EuPathDB this is accomplished with the basket, though less elegantly.
Other systems
Two systems that we did not include in our survey deserve mention. Galaxy (galaxy.psu.edu) (7) is a powerful and popular web-based workflow system that has resemblances to strategies. It is not discussed here because Galaxy is not a database search interface. The Strategies WDK and Galaxy are complementary systems. A user can use the WDK to mine a genome database and when satisfied with a target list of entities, export them to Galaxy for further analysis using a wide array of available tools and resources. The UCSC Genome Browser (genome.ucsc.edu) (26, 27) is also very popular and includes region intersect operations in its Table View, functionality similar to the WDK’s interval logic. We do not include it here because it focuses on sequence intervals rather than integrated functional data, searches and set operations.
Conclusions
The Strategies WDK is a user interface and search system whose development was guided by user input and the recognition that the explosion of data and data types requires a commensurate search tool. While some similar functionality is offered on other sites, the WDK uniquely offers a package of features that makes genomics database mining accessible. It is open source (GNU LGPL), publicly available and has been installed by other groups. With the accelerating growth in diversity and scale of available data sets, the potential for exploiting interrelationships increases dramatically, and we hope this interface will have an impact in bringing ‘genomic scale thinking’ to a broad audience.
Funding
Funding for open access charge: National Institute of Allergy and Infectious Diseases at the National Institutes of Health (Award NO1-AI900038C Contract No. HHSN272200900038C to D.S.R., C.J.S. and J.C.K.).
Conflict of interest. None declared.
Acknowledgments
We wish to thank EuPathDB users and beta-testers, and collaborators interested in adopting the Strategies/WDK system in other bioinformatics applications, for their many helpful comments. We would also like to thank the many members of the EuPathDB team who pushed the WDK to meet their requirements on the EuPathDB sites, as well as developers of early versions of the WDK, Jonathan Crabtree, Thomas Gan and Dave Barkan.



