-
PDF
- Split View
-
Views
-
Cite
Cite
Rosalind J. Cutts, Emanuela Gadaleta, Nicholas R. Lemoine, Claude Chelala, Using BioMart as a framework to manage and query pancreatic cancer data, Database, Volume 2011, 2011, bar024, https://doi.org/10.1093/database/bar024
Close - Share Icon Share
Abstract
We describe the Pancreatic Expression Database (PED), the first cancer database originally designed based on the BioMart infrastructure. The PED portal brings together multidimensional pancreatic cancer data from the literature including genomic, proteomic, miRNA and gene expression profiles. Based on the BioMart 0.7 framework, the database is easily integrated with other BioMart-compliant resources, such as Ensembl and Reactome, to give access to a wide range of annotations alongside detailed experimental conditions. This article is intended to give an overview of PED, describe its data content and work through examples of how to successfully mine and integrate pancreatic cancer data sets and other BioMart resources.
Database URL:http://www.pancreasexpression.org
Project description
In cancer research, advances in technology have resulted in the generation of vast quantities of complex data. Barriers to the effective use of this data include heterogeneity and lack of interoperability between isolated resources (1). We attempted to overcome these obstacles to cancer research by using BioMart (2) to design a generic model for a comprehensive cancer resource. Initially, focusing our efforts on pancreatic cancer, we established the Pancreatic Expression Database (PED) (3–5).
PED is a major resource for the integration and mining of pancreatic cancer literature data. With its data content constantly growing, PED contains the largest collection of pancreatic cancer molecular profiling data sets. Currently, the database includes over 60 000 measurements obtained using a range of omics technologies; including transcriptomics, proteomics, genomics and miRNA. The expansion in data content improved query capabilities and enhanced interoperability have facilitated the systematic study of pancreatic cancer. Although a number of expression data repositories and literature databases exist, PED provides a unique way to integrate and mine data specifically related to pancreatic cancer and cross-query data from other biomart databases and, therefore, allows the community to perform highly detailed queries on facets of pancreatic cancer.
The PED system comprises of tools capable of querying data content either by using simple queries based on an individual premise such as gene expression, or by combining information across multiple data types such as conducting a query addressing both gene expression and copy number data (Figures 1 and 2). By providing the capacity to refine any biological data query according to various criteria, PED provides a resource that allows the pancreatic cancer community to explore and find new relationships among the factors that contribute to the pathogenesis of this disease. The resulting information can be used to elucidate the changes associated with tumourigenesis and the development of resistance to treatment and aid in the development of novel molecular diagnostic tools for the prevention and diagnosis of pancreatic cancer.
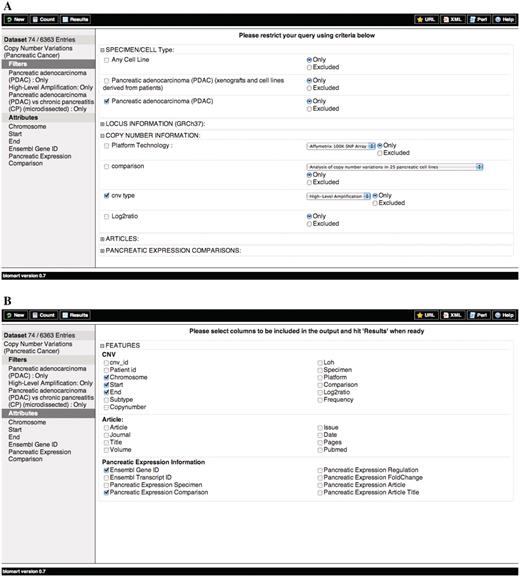
Schematic representation of the querying process for the PED CNV data sets. (A) Filters and (B) Attributes.
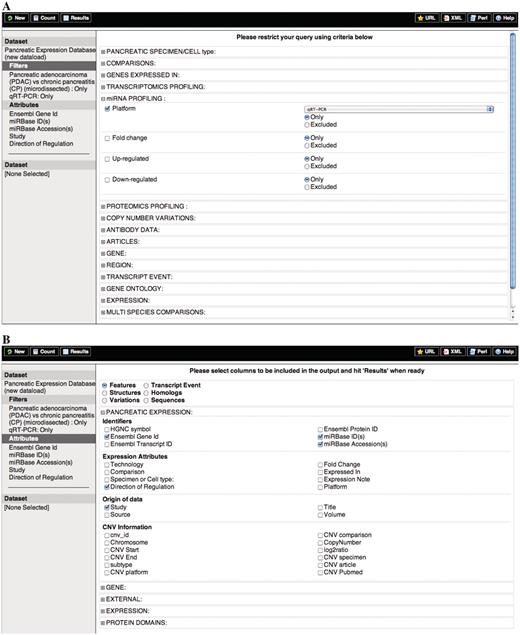
Schematic representation of the querying process for the PED expression data set. (A) Filters and (B) Attributes.
As with all BioMarts, multiple access levels are provided by PED to ensure universal appeal to all members of the research community. The database is freely accessible through a BioMart web-based query interface at: www.pancreasexpression.org. To ensure maximal exposure, PED is also available as a DAS server (6), meaning that it can be used in other resources or browsers such as Ensembl (7). Provision of a data link alerting EntrezGene (8) users of relevant genes in PED is also accessible as a Linkout resource. Moreover, programmatic access is possible through third-party software tools such as R/BioConductor (9), Galaxy (10) and Cytoscape (11). Most importantly, PED is interoperable with the International Cancer Genome Consortium (ICGC) (12), a large-scale collaboration aimed at examining somatically acquired transcriptomic and epigenetic alterations in 50 globally important tumour types or subtypes including pancreatic cancer. The ICGC data portal includes PED annotations on its gene report pages (13).
Data content
The database describes the association of over 6000 DNA copy number alterations; 8000 genes and their 30 000 transcripts and 22 000 proteins; and 279 miRNAs in pancreatic cancer as well as the observed levels of deregulation among a broad range of specimen and experimental types (Table 1). These include healthy/patient tissues and body fluids specimens, cell lines and murine models as well as the provision of information pertaining to any treatments/drugs administered to the samples during the study.
| Data content . | |
|---|---|
| Transcripts | 30 324 |
| Proteins | 22 336 |
| Genes | 8229 |
| miRNA | 279 |
| Genomic copy number alterations: gains and amplifications | 4068 |
| Genomic copy number alterations: losses and deletions | 1420 |
| Genomic copy number alterations: loss of heterozygosity | 875 |
| Data content . | |
|---|---|
| Transcripts | 30 324 |
| Proteins | 22 336 |
| Genes | 8229 |
| miRNA | 279 |
| Genomic copy number alterations: gains and amplifications | 4068 |
| Genomic copy number alterations: losses and deletions | 1420 |
| Genomic copy number alterations: loss of heterozygosity | 875 |
| Data content . | |
|---|---|
| Transcripts | 30 324 |
| Proteins | 22 336 |
| Genes | 8229 |
| miRNA | 279 |
| Genomic copy number alterations: gains and amplifications | 4068 |
| Genomic copy number alterations: losses and deletions | 1420 |
| Genomic copy number alterations: loss of heterozygosity | 875 |
| Data content . | |
|---|---|
| Transcripts | 30 324 |
| Proteins | 22 336 |
| Genes | 8229 |
| miRNA | 279 |
| Genomic copy number alterations: gains and amplifications | 4068 |
| Genomic copy number alterations: losses and deletions | 1420 |
| Genomic copy number alterations: loss of heterozygosity | 875 |
Query examples
PED allows the user to combine complex queries to ask detailed disease-specific questions and combine results with public annotation sources. There are options available for genomics, proteomics, transcriptomics and miRNA profiling, allowing these data types to be queried in isolation or combined (Figures 1 and 2). The interface incorporates the full functionality and data from Ensembl and BioMart for cross-linking different data sets.
To demonstrate the utility of PED, we present several biologically relevant queries that can be performed using the current system.
Query 1: Find genes commonly deregulated in pancreatic cancer precursor lesions, pancreatic intraepithelial neoplasia (PanIN) samples and display gene information, comparison and direction of regulation.
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Specimen: | HGNC symbol |
| PanIN-1b: only | Ensembl Gene Id | |
| PanIN-1b/2: only | Ensembl Transcript ID | |
| PanIN-2: only | Comparison | |
| PanIN-3: only | Direction of regulation |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Specimen: | HGNC symbol |
| PanIN-1b: only | Ensembl Gene Id | |
| PanIN-1b/2: only | Ensembl Transcript ID | |
| PanIN-2: only | Comparison | |
| PanIN-3: only | Direction of regulation |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Specimen: | HGNC symbol |
| PanIN-1b: only | Ensembl Gene Id | |
| PanIN-1b/2: only | Ensembl Transcript ID | |
| PanIN-2: only | Comparison | |
| PanIN-3: only | Direction of regulation |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Specimen: | HGNC symbol |
| PanIN-1b: only | Ensembl Gene Id | |
| PanIN-1b/2: only | Ensembl Transcript ID | |
| PanIN-2: only | Comparison | |
| PanIN-3: only | Direction of regulation |
Predictive biomarkers are vital to cancer research and have been shown to enhance long-term survival for many solid tumours. This data are vital in the identification of asymptomatic, early-stage disease biomarkers. This is especially true for pancreatic cancer, one of the most lethal of solid tumours in which patients tend to be diagnosed in advanced stages of the disease (14).
It is well-established and widely accepted that pancreatic adenocarcinoma (PDAC) progresses from non-invasive pancreatic lesions—pancreatic intraepithelial neoplasia (PanINs) (15). Based on the degree of cellular and nuclear atypia, these precursor neoplasia are delineated as PanIN-1a, PanIN-1b, PanIN-2 and PanIN-3.
The results retrieved in response to Query 1 display genes that have been expressed in all the PanIN collection included in the database. The results table will also provide links to the original studies. In this instance, the deregulation of the S100P gene is highlighted as an early event in the development of pancreatic cancer. This is in accordance with previous published findings (16,17).
Query 2: Find genes differentially expressed in the serum of pancreatic cancer patients when compared to the serum of patients with benign pancreatic diseases (chronic pancreatitis and pancreatic pseudocyst). Find associated pathways via query integration with Reactome. Display gene and protein information, experimental details and pathway information.
| Data sets . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Comparison: | HGNC symbol |
| Pancreatic cancer versus benign pancreatic disease (CP and pancreatic pseudocyst) (serum) | Ensembl Gene Id | |
| Limit output to: pancreatic cancer patients/benign pancreatic disease (chronic pancreatitis and pancreatic pseudocyst) (serum) | Ensembl Protein ID | |
| Comparison | ||
| Direction of Regulation | ||
| Fold change | ||
| Platform | ||
| Reactome [pathway] | Species: Homo sapiens | Pathway name |
| Pathway DB ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Comparison: | HGNC symbol |
| Pancreatic cancer versus benign pancreatic disease (CP and pancreatic pseudocyst) (serum) | Ensembl Gene Id | |
| Limit output to: pancreatic cancer patients/benign pancreatic disease (chronic pancreatitis and pancreatic pseudocyst) (serum) | Ensembl Protein ID | |
| Comparison | ||
| Direction of Regulation | ||
| Fold change | ||
| Platform | ||
| Reactome [pathway] | Species: Homo sapiens | Pathway name |
| Pathway DB ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Comparison: | HGNC symbol |
| Pancreatic cancer versus benign pancreatic disease (CP and pancreatic pseudocyst) (serum) | Ensembl Gene Id | |
| Limit output to: pancreatic cancer patients/benign pancreatic disease (chronic pancreatitis and pancreatic pseudocyst) (serum) | Ensembl Protein ID | |
| Comparison | ||
| Direction of Regulation | ||
| Fold change | ||
| Platform | ||
| Reactome [pathway] | Species: Homo sapiens | Pathway name |
| Pathway DB ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Comparison: | HGNC symbol |
| Pancreatic cancer versus benign pancreatic disease (CP and pancreatic pseudocyst) (serum) | Ensembl Gene Id | |
| Limit output to: pancreatic cancer patients/benign pancreatic disease (chronic pancreatitis and pancreatic pseudocyst) (serum) | Ensembl Protein ID | |
| Comparison | ||
| Direction of Regulation | ||
| Fold change | ||
| Platform | ||
| Reactome [pathway] | Species: Homo sapiens | Pathway name |
| Pathway DB ID |
Determination of reliable non-invasive biomarkers, such as those present in serum, are important when attempting to avoid surgical intervention and limit any physiological and psychological stress on the patient. PED not only allows users to query profiles derived from tissues but also those from media such as serum, plasma and urine. In addition, linking of the PED resource to Reactome (18) enables both the identification of potential genes specific to pancreatic cancer for biomarker discovery and visualization of the affected pathways.
Query 3: Find DNA copy number high-level amplifications in PDAC samples that also contain genes differentially expressed in PDAC versus chronic pancreatitis (CP) and display copy number information, gene information and differential expression experimental details (Figure 1).
| Data set . | Filters . | Attributes . |
|---|---|---|
| Copy number variations (pancreatic cancer) | Specimen/cell type: pancreatic adenocarcinoma (PDAC): only | Chromosome |
| Copy number information: high-level amplification: only | Start | |
| Pancreatic expression comparisons: pancreatic adenocarcinoma (PDAC) versus chronic pancreatitis (CP) (microdissected): only | End | |
| Ensembl Gene ID | ||
| Pancreatic expression comparison |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Copy number variations (pancreatic cancer) | Specimen/cell type: pancreatic adenocarcinoma (PDAC): only | Chromosome |
| Copy number information: high-level amplification: only | Start | |
| Pancreatic expression comparisons: pancreatic adenocarcinoma (PDAC) versus chronic pancreatitis (CP) (microdissected): only | End | |
| Ensembl Gene ID | ||
| Pancreatic expression comparison |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Copy number variations (pancreatic cancer) | Specimen/cell type: pancreatic adenocarcinoma (PDAC): only | Chromosome |
| Copy number information: high-level amplification: only | Start | |
| Pancreatic expression comparisons: pancreatic adenocarcinoma (PDAC) versus chronic pancreatitis (CP) (microdissected): only | End | |
| Ensembl Gene ID | ||
| Pancreatic expression comparison |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Copy number variations (pancreatic cancer) | Specimen/cell type: pancreatic adenocarcinoma (PDAC): only | Chromosome |
| Copy number information: high-level amplification: only | Start | |
| Pancreatic expression comparisons: pancreatic adenocarcinoma (PDAC) versus chronic pancreatitis (CP) (microdissected): only | End | |
| Ensembl Gene ID | ||
| Pancreatic expression comparison |
The query above shows a simple way to integrate multi-dimensional data by combining data on copy number variations with results from differential expression. This will give an overview of genomic or transcriptomic changes of any chromosome(s) or chromosomal region(s) in the selected sample(s) as determined by a variety of platforms stored in PED. By overlaying the information on transcriptional deregulation from expression arrays and gene content with copy number variations from genomic arrays, one can quickly highlight the commonly affected regions in the studied patient population and the impact of the copy number aberrations on gene expression patterns. For example, by looking for high-level amplifications and genes up-regulated in PDAC when compared to chronic pancreatitis, it is possible to highlight potential oncogenes.
Query 4: Find miRNAs differentially expressed in PDAC versus CP whose expression has been confirmed by RT–PCR techniques and display miRNA attributes and study information (Figure 2).
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Gene: Gene type: miRNA miRNA Profiling: Platform: qRT-PCR Comparison: PDAC versus CP (microdissected) | Ensembl Gene Id miRBase ID(s) miRBase Accession(s) Study Comparison Direction of regulation |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Gene: Gene type: miRNA miRNA Profiling: Platform: qRT-PCR Comparison: PDAC versus CP (microdissected) | Ensembl Gene Id miRBase ID(s) miRBase Accession(s) Study Comparison Direction of regulation |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Gene: Gene type: miRNA miRNA Profiling: Platform: qRT-PCR Comparison: PDAC versus CP (microdissected) | Ensembl Gene Id miRBase ID(s) miRBase Accession(s) Study Comparison Direction of regulation |
| Data set . | Filters . | Attributes . |
|---|---|---|
| Pancreatic Expression Database | Gene: Gene type: miRNA miRNA Profiling: Platform: qRT-PCR Comparison: PDAC versus CP (microdissected) | Ensembl Gene Id miRBase ID(s) miRBase Accession(s) Study Comparison Direction of regulation |
miRNAs bind to target sites in the 3′-UTR region of mRNA and act as repressors of protein-coding genes or activators of RNA degradation. Aberrant expression of miRNAs involved in pancreatic cancer can be easily retrieved from PED.
Discussion and future directions
PED has become well-established in the pancreatic cancer community as a key resource for mining relevant literature information. Recent updates have added in different information types including copy number variations from pancreatic cancer samples and other expression experiments such as proteomics and miRNA.
The successful development and implementation of PED fills the urgent requirement of the pancreatic cancer community for resources capable of integrating the overflowing influx of data generated by novel high-throughput technologies.
The architectural flexibility of PED BioMart-based schema means that it can be easily extended to encompass additional malignant and non-malignant diseases and has been used as a prototype for other malignant diseases such as breast cancer (http://bioinformatics.breastcancertissuebank.org).
The use of BioMart as a framework facilitates interoperability with other cancer resources and enables users to cross-query data from a number of relevant resources rather than being limited to a single database. The International Cancer Genome Consortium uses BioMart technology to share data and make it publicly available. Data from PED is automatically cross-queried from the ICGC (see ICGC paper in this issue) and can be queried with COSMIC data (19) via the BioMart framework. This allows direct cross-comparison of experimental findings generated from the two ICGC pancreatic cancer projects (Australia and Canada) with literature-derived information from PED.
Plans for the database include expanding to include reanalysed differential expression data and methods to enable users to assess the quality of the information added to the database. There are also plans to improve the graphical data view, especially for genomic information.
Funding
Cancer Research UK (programme grant C355/A6253) and FW6 EU project MolDiag-Paca; Breast Cancer Campaign (to R.J.C.). Funding for open access charge: Cancer Research UK (programme grant C355/A6253).
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank pancreatic cancer scientists who have suggested or contributed literature data for the database.



