-
PDF
- Split View
-
Views
-
Cite
Cite
Chih-Hsuan Wei, Bethany R. Harris, Donghui Li, Tanya Z. Berardini, Eva Huala, Hung-Yu Kao, Zhiyong Lu, Accelerating literature curation with text-mining tools: a case study of using PubTator to curate genes in PubMed abstracts, Database, Volume 2012, 2012, bas041, https://doi.org/10.1093/database/bas041
Close - Share Icon Share
Abstract
Today’s biomedical research has become heavily dependent on access to the biological knowledge encoded in expert curated biological databases. As the volume of biological literature grows rapidly, it becomes increasingly difficult for biocurators to keep up with the literature because manual curation is an expensive and time-consuming endeavour. Past research has suggested that computer-assisted curation can improve efficiency, but few text-mining systems have been formally evaluated in this regard. Through participation in the interactive text-mining track of the BioCreative 2012 workshop, we developed PubTator, a PubMed-like system that assists with two specific human curation tasks: document triage and bioconcept annotation. On the basis of evaluation results from two external user groups, we find that the accuracy of PubTator-assisted curation is comparable with that of manual curation and that PubTator can significantly increase human curatorial speed. These encouraging findings warrant further investigation with a larger number of publications to be annotated.
Database URL:http://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/PubTator/
Introduction
In order for manual curation to keep up with the rapid growth of the biomedical literature, past research (1–3) has suggested taking advantage of the research and development of biomedical text-mining and natural language processing. However, despite multiple attempts from the text-mining community (4–8), to date, still few existing text-mining tools have been successfully integrated into production systems for literature curation (9,10).
Textpresso (11), an information extracting and processing system for biological literature, is one such exception. According to the previous study (9), a key ingredient to its success is the fact that Textpresso grew directly out of the curation community. More specifically, Textpresso was developed in collaboration with WormBase (12) for its specific curation tasks. Thus, from its initial development to the final deployment into production, the Textpresso tool developers worked closely with the WormBase curators. The lack of such close working relationships between tool developers and end users is one of the limiting factors in advancing computer-assisted literature curation.
To promote interactions between the biocuration and text-mining communities, an interactive text-mining track (hereafter, ‘Track III’) was held in the BioCreative (Critical Assessment of Information Extraction systems in Biology) 2012 workshop (13). Track III provides volunteer biocurators the chance to participate in a user study of a chosen system and text-mining teams the opportunity to collect interactive data. Teams define a curation task and provide a gold-standard biomedical literature corpus, while the curators are responsible for curating the desired data from the corpus, performing half of the work manually and half through interaction with the system.
The Track III challenge provides valuable evaluation of the participating text-mining systems. While performing the tasks, biocurators track time so that research teams can then compute time-on-task and efficiency of their systems’ use. PubTator (14) was formally evaluated before the BioCreative 2012 workshop by two external user groups: the Arabidopsis Information Resource (TAIR) and the National Library of Medicine (NLM). (The NLM evaluator was from Library Operation, external to the PubTator development team.) TAIR maintains a database of genetic and molecular biology data for the model higher plant Arabidopsis thaliana (15) and has been curating information from the literature for >10 years. Results from both manual and assisted curation are compared against the gold standard for measuring annotation quality. Biocurators also complete a post-study survey consisting of questions about task completion, which provides research teams with user feedback on the usability of the system.
Materials and methods
Evaluation tasks for PubTator
As mentioned earlier, PubTator was formally evaluated before the BioCreative 2012 workshop by two external user groups: TAIR and NLM. Specifically, a TAIR curator used PubTator for both document triage and bioconcept annotation tasks, whereas an NLM curator evaluated PubTator only for bioconcept annotation. Although PubTator may be used for the annotation of a variety of bioconcepts, both of our proposed tasks focused on gene indexing, a task that is central to all model organism databases and many other curation groups. The PubTator environment was also appropriately tailored for each user group, providing customized versions that most suited the biocurators’ respective tasks.
Following the BioCreative 2012 Track III guidelines, for each evaluation, we asked a human curator to process a total of 50 documents in two settings: curate one collection of 25 PubMed abstracts manually and the other set of 25 abstracts with the use of PubTator. Manual processing involved curation with the use of the PubMed environment and storing the results in spreadsheets. Using PubTator, curators could accept, edit or reject output provided by the system and then store the validated information in the system. For the PubTator-assisted gene indexing, the biocurators were reviewing machine-tagged pre-annotations of gene names and accepting, adding to or adjusting the PubTator output. Manual gene indexing consisted of looking up the relevant gene identifier in the appropriate online resource (see below).
As shown in Table 1, the two test collections were sampled from past curated data provided by TAIR and NLM. We ensured that the gold-standard annotations were created by someone separate from the biocurator participating in the PubTator evaluation.
| Group . | Gold standard (50 abstracts) . | Curation tasks . |
|---|---|---|
| NLM | Sampled from the 151 gene indexing assistant test collection | Gene indexing (mention level) |
| TAIR | Sampled from all the papers reviewed by the TAIR group in December 2011 | Gene indexing (document level) |
| Document triage |
| Group . | Gold standard (50 abstracts) . | Curation tasks . |
|---|---|---|
| NLM | Sampled from the 151 gene indexing assistant test collection | Gene indexing (mention level) |
| TAIR | Sampled from all the papers reviewed by the TAIR group in December 2011 | Gene indexing (document level) |
| Document triage |
| Group . | Gold standard (50 abstracts) . | Curation tasks . |
|---|---|---|
| NLM | Sampled from the 151 gene indexing assistant test collection | Gene indexing (mention level) |
| TAIR | Sampled from all the papers reviewed by the TAIR group in December 2011 | Gene indexing (document level) |
| Document triage |
| Group . | Gold standard (50 abstracts) . | Curation tasks . |
|---|---|---|
| NLM | Sampled from the 151 gene indexing assistant test collection | Gene indexing (mention level) |
| TAIR | Sampled from all the papers reviewed by the TAIR group in December 2011 | Gene indexing (document level) |
| Document triage |
The NLM test collection was taken from the existing Gene Indexing Assistant (GIA) test collection (http://ii.nlm.nih.gov/TestCollections/index.shtml#GIA), which is a corpus consisting of manually annotated MEDLINE citations, randomly chosen from human genetics journals published between 2002 and 2011. Explicit mentions of genes and gene products were normalized to the appropriate National Center for Biotechnology Information (NCBI) Entrez Gene identifier. The sole annotator of the GIA corpus provided annotation guidance to the NLM volunteer biocurator.
The TAIR test set is different from the NLM counterpart in two major aspects. First, their gene annotation is different: for each abstract in the NLM test set, every gene name mention is annotated and normalized to an Entrez Gene ID. Conversely, only unique gene identifiers are kept for each abstract in the TAIR set. Moreover, in lieu of using Entrez Gene, TAIR uses its own nomenclature for Arabidopsis genes (which was accommodated by PubTator through customization for TAIR tasks). Second, each TAIR abstract is also assigned with an additional label that indicates whether the paper qualifies for full curation.
We were primarily interested in how using PubTator affected the speed and accuracy of the biocurators’ work. Participants were asked to install a Firefox Web browser add-on to record time-on-task and user interactions with the system. They recorded their own time for the manual tasks. Precision and recall measures for the manual and assisted curation sets were benchmarked against the provided gold-standard annotations. Finally, biocurators were asked to provide feedback on task completion and system usability via a workshop-provided online survey.
Evaluation metrics
We first compared the biocurator’s curation results with the gold standard so that we are able to see whether a curator’s accuracy changes with and without PubTator. For this purpose, we used the traditional precision, recall and F-measure metrics (16). More importantly, we evaluated PubTator’s ability to improve curation efficiency. Specifically, we compared the average time (in seconds) needed to complete curating an abstract with and without the use of PubTator.
PubTator design
PubTator (14) was developed based on a prototype system that was previously used at the NCBI for various manual curation projects, such as annotating disease mentions in PubMed abstracts (17,18). We significantly extended our previous system in developing PubTator. First, relevance ranking and concept highlighting were added to ease the task of document triage. Second, state-of-the-art named entity recognition tools [e.g. competition-winning gene normalization systems (19,20) in BioCreative III (5)] and our newly developed species recognition tool SR4GN (21) were integrated to pre-tag bioconcepts of interest, as a way to facilitate the task of gene annotation. Third, PubTator was developed to have a look and feel similar to PubMed, thus minimizing the learning effort required for new users. Furthermore, a standard PubMed search option is made available in PubTator, which would allow our users to make a hassle-free move of their saved PubMed queries (a common practice for curators doing document triage) into this new curation system. Finally, by taking advantage of pre-tagging bioconcepts, PubTator also allows its users to perform semantic search besides the traditional keyword-based search, a novel feature not available in PubMed.
Results and discussion
Evaluation data sets
The gold-standard corpora and associated characteristics are described below (Table 2). For each user group’s task, two sets of 25 abstracts (50 total) that had similar characteristics were selected out of the entire gold-standard corpora. The text-mining team ensured that the two test sets to be curated with and without the use of PubTator similar to one another with respect to the number of gene mentions (according to the gold standard). The unannotated copies of the corpora were then sent to the volunteer biocurators for manual annotation.
Table 2. The statistics of testing corpora for PubTator evaluation
| Gold standard . | PubMed set (25 docs) . | PubTator set (25 docs) . |
|---|---|---|
| NLM—gene indexing | 188 Gene mentions | 172 Gene mentions |
| TAIR—gene indexing | 44 Gene identifiers | 29 Gene identifiers |
| TAIR—document triage | 13 Relevant articles | 11 Relevant articles |
| Gold standard . | PubMed set (25 docs) . | PubTator set (25 docs) . |
|---|---|---|
| NLM—gene indexing | 188 Gene mentions | 172 Gene mentions |
| TAIR—gene indexing | 44 Gene identifiers | 29 Gene identifiers |
| TAIR—document triage | 13 Relevant articles | 11 Relevant articles |
Table 2. The statistics of testing corpora for PubTator evaluation
| Gold standard . | PubMed set (25 docs) . | PubTator set (25 docs) . |
|---|---|---|
| NLM—gene indexing | 188 Gene mentions | 172 Gene mentions |
| TAIR—gene indexing | 44 Gene identifiers | 29 Gene identifiers |
| TAIR—document triage | 13 Relevant articles | 11 Relevant articles |
| Gold standard . | PubMed set (25 docs) . | PubTator set (25 docs) . |
|---|---|---|
| NLM—gene indexing | 188 Gene mentions | 172 Gene mentions |
| TAIR—gene indexing | 44 Gene identifiers | 29 Gene identifiers |
| TAIR—document triage | 13 Relevant articles | 11 Relevant articles |
Comparison of curation accuracy with versus without PubTator assistance
As we can see in Figure 1, the human curator accuracies are generally high for all NLM and TAIR tasks (over 80% in F-measure), suggesting that the testing experiments were performed rigorously. In fact, with the aid of PubTator, all figures indicate that a human curator can curate literature slightly more accurately with the assistance of a text-mining tool than doing this completely by hand, although not statistically significant according to Fisher’s randomization test (22). Precision and recall measures indicated that annotations were quite similar to those of the gold standard corpus.
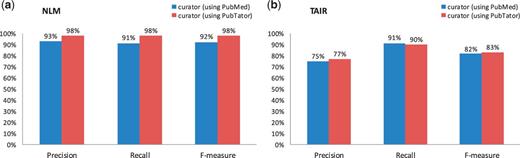
Comparison of human curation accuracy for the gene indexing task by using PubMed versus PubTator. (a) NLM mention-level results. (b) TAIR document-level results.
Despite high accuracy of all tasks, the human curator results do not completely match the gold standard. The imperfect F-measures in both figures might be caused by potential changes to the curation guidelines and individual differences between curators. The difference in the F-measures of Figure 1b versus Figure 2 suggests that the gene indexing task is more difficult for human curators than the document triage task, for which the only measure that is <90% is the recall (85%) in the PubTator set. Our further analysis shows that this was essentially due to the miss of two relevant papers (of 13 totally) by the TAIR human curator. One misclassified article (23) contains multiple species (Human, Drosophila, Caenorhabditis elegans and Arabidopsis) in the abstract, whereas the other (24) mentions none. Only their full text makes it clear that both papers are relevant for TAIR curation. However, only abstracts were used for making decisions in the current experiment.
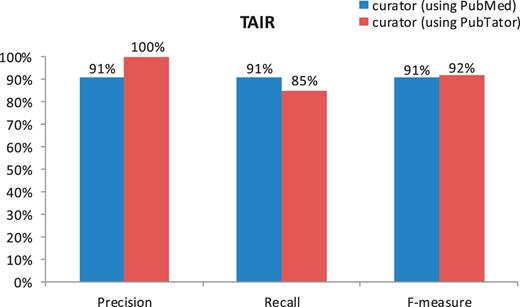
Comparison of human curation accuracy for the document triage task by using PubMed versus PubTator (TAIR).
Comparison of curation efficiency with versus without PubTator assistance
Figure 3a shows that, on average, the NLM curator needed 326 s to curate an abstract completely by hand. With PubTator, the required time decreases to 190 s, a 42% improvement in curation efficiency.
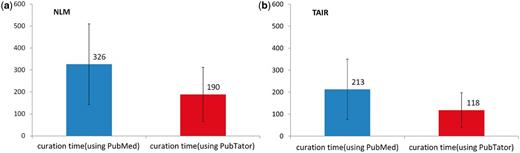
Comparison of human curation speed for the gene indexing task by using PubMed versus PubTator. The black bars represent the standard deviation of curation time. (a) NLM results. (b) TAIR results.
In comparison, the TAIR curation task is considerably less time-consuming, as here only unique gene IDs were required as opposed to each gene mention. Therefore, as shown in Figure 3b, the TAIR curator averaged 213 s to manually curate each abstract, while only taking an average of 118 s to process PubTator-assisted annotations, resulting in a 45% increase in efficiency (an improvement similar to the NLM task result).
Changes to PubTator based on user feedback
During and after the pre-workshop evaluation, we received useful feedback from our users and made several improvements to PubTator accordingly. Some of the notable adjustments include the creation of a PubTator collection management feature and new functionalities for removing and copying an existing annotation.
To help users manage multiple annotation projects in PubTator, we developed a new document management system by which a user can create a document collection for each project and use a different annotation environment for each collection. For instance, a user can select that only pre-annotated gene results be shown in a particular collection by deselecting the results of the three other bioconcepts. In fact, besides the ability to selectively display the four default bioconcepts, users can also create their own concepts of interest inside their personal collections.
It is common that a named entity, such as a gene name, is mentioned multiple times in an abstract. Thus, instead of requiring users to annotate the same mention at its every occurrence, we implemented a new ‘copy’ function such that by a single click associated with an existing mention, all of its occurrences elsewhere in the same abstract will be automatically captured. In a similar fashion, we made the removal of an existing annotation straightforward, using a single keystroke.
Conclusions and future work
On the basis of user evaluation results from two independent curation groups, we conclude that PubTator-assisted curation can significantly improve curation efficiency by over 40% without any loss in the quality of final annotation results. These encouraging findings warrant further investigation with a larger number of publications to be annotated. Furthermore, it is worth comparing actual gains using PubTator versus curators’ existing working environment in future research. For instance, despite the fact that our baseline setting (using PubMed and spreadsheet) was the actual environment for NLM curators, TAIR already has its own curation tool. Such comparisons are more meaningful for individual groups to select computer assistant tools.
Despite its promising results in BioCreative 2012 Track III, PubTator has several limitations. First, PubTator currently pre-annotates only four named entities (i.e. gene, disease, chemical and species) by design. Many other important bioconcepts (e.g. Gene Ontology terms) are missing from its pre-tagged results. Second, PubTator currently only works for the bioconcept annotation and document triage tasks. It cannot be used to identify relationships between bioconcepts, such as protein–protein interaction. The final limitation of PubTator is its ability in handling full text. Only PubMed abstracts are now supported for annotation in PubTator. We plan to address the aforementioned issues in our future work.
Funding
Intramural Research Program of the NIH, National Library of Medicine (to C.W. and Z.L.); National Library of Medicine and administered by the Oak Ridge Institute for Science and Education (to B.H.); National Science Foundation (DBI-0850219 to TAIR); TAIR sponsors (http://www.arabidopsis.org/doc/about/tair_sponsors/413). Funding for open access charge: National Institutes of Health, National Library of Medicine.
Conflict of interest. None declared.
Acknowledgements
We are grateful to Larry Smith, Donald Comeau and Rezarta Islamaj Doğan for building the prototype annotation system. We also thank W. John Wilbur and Sun Kim for helpful discussion. Finally, we thank Caitlin Sticco for providing the gold-standard corpus from the NLM Gene Indexing Assistant test collection for use in the NLM component of the BioCreative 2012 Track III task.



