-
PDF
- Split View
-
Views
-
Cite
Cite
Ankur Gautam, Harinder Singh, Atul Tyagi, Kumardeep Chaudhary, Rahul Kumar, Pallavi Kapoor, G. P. S. Raghava, CPPsite: a curated database of cell penetrating peptides, Database, Volume 2012, 2012, bas015, https://doi.org/10.1093/database/bas015
Close - Share Icon Share
Abstract
Delivering drug molecules into the cell is one of the major challenges in the process of drug development. In past, cell penetrating peptides have been successfully used for delivering a wide variety of therapeutic molecules into various types of cells for the treatment of multiple diseases. These peptides have unique ability to gain access to the interior of almost any type of cell. Due to the huge therapeutic applications of CPPs, we have built a comprehensive database ‘CPPsite’, of cell penetrating peptides, where information is compiled from the literature and patents. CPPsite is a manually curated database of experimentally validated 843 CPPs. Each entry provides information of a peptide that includes ID, PubMed ID, peptide name, peptide sequence, chirality, origin, nature of peptide, sub-cellular localization, uptake efficiency, uptake mechanism, hydrophobicity, amino acid frequency and composition, etc. A wide range of user-friendly tools have been incorporated in this database like searching, browsing, analyzing, mapping tools. In addition, we have derived various types of information from these peptide sequences that include secondary/tertiary structure, amino acid composition and physicochemical properties of peptides. This database will be very useful for developing models for predicting effective cell penetrating peptides.
Database URL:http://crdd.osdd.net/raghava/cppsite/.
Introduction
Advances in proteomics and high-throughput assays have led to the discovery of a large number of highly potent therapeutic molecules. However, most of these molecules do not reach the clinical trial stage due to their poor delivery and low bioavailability. The plasma membrane of eukaryotic cell is selectively permeable for exogenous molecules and large therapeutic molecules like DNA, proteins and many drugs are impermeable due to their size or biochemical properties. The intracellular delivery of these molecules has always been proven a major challenge for scientific community. In order to overcome these problems, many drug delivery techniques (viral and non-viral) have been developed over the years (1, 2). Cell penetrating peptides (CPPs) based method is one of the powerful techniques used for delivering therapeutics/ drugs (3–6). It has numerous advantages over the other delivery methods like versatility, high efficiency, low toxicity.
Most of the CPPs have following features, short size (10–30 amino acids), water soluble, often consists of basic amino acids (arginine and lysine), and are mostly cationic or amphipathic in nature (7–9). On the basis of their origin, CPPs can be divided into three major categories; (i) protein derived peptides (e.g. Tat, penetratin), (ii) chimeric peptides (e.g. transportan) and (iii) synthetic peptides (10). These peptides have been used to deliver various types of cargoes such as proteins, peptide, siRNA, liposomes, nanoparticles and drugs (11). In past, CPPs have been used in various clinical applications particularly in cancer therapy (6, 12). Recently, tumor homing CPPs (13) and activatable CPPs (14) have been designed and used in the treatment and diagnosis of cancer in vitro and in vivo. In addition, CPPs have also been used in vaccine designing (15). Tat peptide has been used for delivery of tumor associated cancer antigen in antigen presenting cells (16, 17).
In summary, CPPs have the tremendous potential, particularly in the field of therapeutics. Thus, there is a need to compile these peptides from literature in order to understand properties of these peptides. To the best of author’s knowledge, no database or resource provides comprehensive information about CPPs. In this study, we have made a systematic attempt to collect and compile information on CPPs from published literature, patents and other resources. We hope this database will be very useful for researchers working in the field of therapeutics.
Material and methods
Data collection and compilation
CPPs were collected and compiled from various resources. It includes around 500 research articles and 50 patents. These research articles and patents were extracted from literature/patent databases using a combination of keywords like cell-penetrating peptides, membrane permeable peptides, protein transduction domain and membrane transporting peptides. After careful readings of these articles, we have scrutinized 83 research articles and 20 patents. From these articles and patents, only experimentally validated CPP sequences and other relevant experimental information like end modification, uptake efficiency, uptake mechanism, sub-cellular localization and cell lines used have been extracted and compiled. We have made multiple entries of the CPPs, if similar peptides have been tested against different cell types or similar peptides have different end modifications. Therefore, total entries in CPPsite are 843, but unique CPP sequences are 741.
Database architecture and web interface
It is built on an Apache HTTP Server 2.0.59 with MySQL server. The front-end was developed using HTML, PHP, JAVA script and the back-end was developed using MySQL, a relational database management system. All common gateway interface (CGI) and database interfacing scripts were written in the PHP and PERL programming language. Apache, MySQL and PHP technology were preferred as they were platform independent and open-source software. The architecture of CPPsite database is shown in Figure 1.
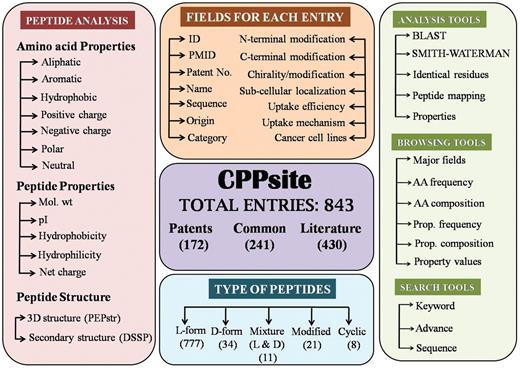
Organization of data
CPPsite is a manually curated database which provides comprehensive information about each CPP. Data for each peptide can be categorized as primary (e.g. peptide sequence, PubMed ID (PMID) and experimental details) and secondary data (e.g. secondary/tertiary structure, amino acid composition, frequency and physicochemical properties). Each peptide is assigned a unique entry number, and information is divided into different tables. Each table provides unique information.
Primary data
Primary data contains general information of CPPs. All the information is extracted manually from various resources, mainly from publications and patents. As shown in the Figure 1, database provides comprehensive information on each peptide using more than 15 types of fields. Main fields are described as follows: (i) Name: it represents the name of the peptide used in literature; (ii) Sequence: it contains amino acid sequence of CPP; (iii) Nature: it contains the nature (e.g. cationic) of peptide; (iv) Family: it represents the class of peptide (e.g. protein derived); (v) uptake efficiency: It provides relative uptake efficiency of CPP; (vi) uptake mechanism: it provides the mechanism of internalization of CPP; (vii) end modification: it represents the modification (e.g. FITC labeling/ amidation) at N-and C-terminus of peptide; (viii) Cell lines: it represents the cell lines used to validate the CPP.
Secondary data
In the past, it has been shown that internalization of CPP depends on its structure (18) and nature of peptide. Therefore, understanding of tertiary structure of these peptides is a prerequisite. Since most of the peptide structures are not available in Protein Data Bank (PDB) and existing methods for predicting tertiary structure of protein are unsuitable for predicting tertiary structure of peptides, we have predicted tertiary structure of CPPs using software PEPstr (19). PEPstr is a state of art method for predicting structure of bioactive peptides. It is a de-novo method which predicts beta-turns in a given peptide sequence followed by secondary structure prediction by PSIPRED and allocation of side-chain torsion angles by Dunbrack Library. Energy minimization of the final model is done using AMBER Version 6. In addition, secondary structures of these peptides have also been predicted from predicted tertiary structure using software DSSP (20). In our database, we maintain both secondary and tertiary structure of each peptide in PDB format.
In order to understand the nature of the peptide, it is imperative to know the properties (pI, molecular weight, amino acid frequency, hydrophobicity, charge, etc.) of each peptide. Therefore, in-house PERL scripts have been used for computing frequency and composition of each type of amino acid residue in all the peptides. This information is very useful to understand which residues/motifs are preferred in CPPs. Apart from amino acid frequency and composition; users may also want to know which types of residues are preferred like charged, polar, hydrophobic, etc. Therefore, we have computed frequency and composition of each class (aromatic, aliphatic, positive charge, negative charge, neutral, polar and hydrophobic) of residues. This information is very useful to know which peptides are dominated by positively charged residues. We have also computed overall physicochemical properties (hydrophobicity, hydrophilicity, molecular weight, iso-electric point and net charge) of each peptide (21, 22). All above information is stored in the database as secondary data for analysis and browsing of peptides based on their properties.
Implementation of tools
Currently, CPPsite contains 843 peptides, which have been studied against almost 85 different cell lines. Experimental details, PMID and patents information are also given for every peptide. Many user-friendly tools/interfaces have been integrated in CPPsite for extraction and analysis of database. Following are the main tools provided with the CPPsite database:
Search tools
CPPsite provides three search tools: simple search, advanced search and peptide search. Simple search option allows users to perform the search on any field of the database like PubMed ID, CPP name, CPP sequence, origin, nature of CPP (cationic and amphipathic), uptake mechanism, etc. This option also allows displaying any or all fields of databases for selected searched records. Advanced search facility allows users to perform multiple queries at a time. Under advanced search option, users can add any number of queries for performing a search in the database. In addition, it allows users to select conditions (e.g. AND & OR) between queries. In summary, advanced search option allows users to perform any type of data retrieval from CPPsite. Peptide search option provides an option for searching a peptide sequence in the database. It allows two types of peptide search, (i) Containing peptide: it is for searching user defined peptide sequence in CPPs, and (ii) Exact search: it allows users to search CPPs, which are 100 percent identical to user’s peptide.
Browsing tools
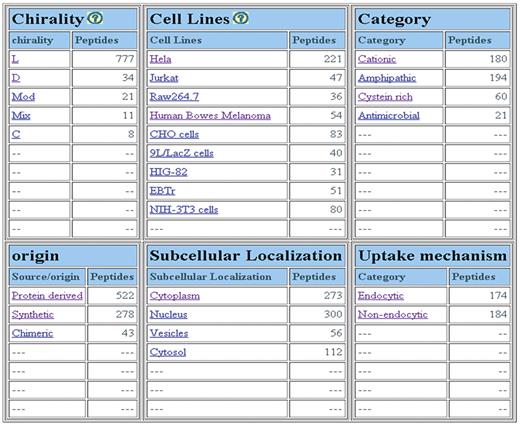
We have designed powerful browsing facility that allows a user to browse data on major fields that includes (i) chirality of peptides; (ii) nature of peptides; (iii) family of peptides; (iv) cell lines; (v) uptake mechanism and (iv) sub-cellular localization. Chirality field includes five types of peptides: peptides having all l-amino acids; all d-amino acids; both l- & d-amino acids (mixed), and non-natural amino acids (modified). Cyclic (C) peptides have also been included in this field. As shown in Figure 2, it provides a number of peptides for different types of CPPs, uptake mechanism, sub-cellular localization and cell lines. It allows users to browse on secondary data like (i) amino acid frequency, (ii) amino acid composition, (iii) physicochemical property frequency, (iv) physicochemical property composition and (v) physicochemical property value (21, 22).
Retrieval and visualization of structures
In this database, predicted secondary and tertiary structure of each peptide has been stored. Three interfaces have been developed to extract structural information of CPPs. First interface allows users to browse peptides based on their secondary structure composition (composition H-helix, E-β strand, T-turn and C-coil). Second interface allows users to search secondary structure segment in secondary structure of CPPs.
Third interface is designed to browse tertiary structure of CPPs. We have integrated Jmol (http://www.jmol.org) to CPPsite that allows users to visualize 3D structure of CPPs.
Web-based tools
A number of web-based tools have been integrated in this database to facilitate further analysis of peptides. A brief description of these tools is as follows:
Blast search
We have integrated BLAST search tool (23) that allows users to perform similarity based search against CPPs. This option allows users to submit one or more peptide sequences in FASTA format for performing BLAST search against CPPs.
Smith–Waterman algorithm
In order to handle similarity search effectively in case of small peptides, we have integrated Smith–Waterman algorithm (24). This option allows users to search CPPs in the database that are similar to their peptides. Users can submit multiple peptide sequences in FASTA format.
Identical residues
In addition to BLAST and Smith–Waterman algorithm, a simple algorithm has been integrated called identical residues. It takes user defined peptide sequence and aligns it with each CPP using overlapping approach (alignment without gap). It displays query and target sequence with a number of identical residues in two peptides.
Peptide mapping
It allows users to map CPPs on their peptide sequence. Users may submit protein or polypeptide sequence on this page to identify segments that are identical to peptides in CPPsite.
Results
CPPsite is a unique collection of experimentally validated 843 CPPs and their derivatives in which 430 CPPs have been collected from research publications, 172 CPPs from patents and 241 CPPs from both research papers and patents. The important feature of CPPsite is that it covers all possible types of CPPs that consists of l-amino acids (777 peptides), D-amino acids (34 peptides), both l & d amino acids (11 peptides) and non-natural amino acids (21 peptides). Few cyclic peptides (8 peptides) have also been incorporated (Figure 2). Most of the peptides (522) are protein derived, 278 peptides are designed/synthetic and 43 peptides are chimeric in nature. CPPsite contains 180 cationic (Arg rich) and 194 amphipathic peptides. Few of them are antimicrobial (21 peptides) in nature (Figure 2).
CPPs have been validated for their cell penetration capability on variety of cell types in different conditions. In CPPsite, CPPs have been found to be internalized into more than 80 types of cell lines. As shown in Figure 3A, most of the CPPs have been tested against HeLa cells (221 peptides), CHO (83 peptides), Human bowes melanoma cell (54 peptides), Jurkat (47 peptides), EBTr (51 peptides) and NIN-3T3 cells (80 peptides). CPPs have shown different uptake mechanisms, which depend mainly on the size (cargo i.e. peptides, proteins, fluorophores etc), temperature and cell types (5, 25, 26). In many cases, similar peptides have shown different uptake mechanism at different temperatures and at different concentrations (5, 25). In CPPsite, 174 peptides displayed endocytic uptake mechanism, while 184 peptides showed non-endocytic uptake mechanism (Figure 3B). In non-endocytic route, peptides have been found to be localized mainly into the cytoplasm and nucleus, while in case of endocytic uptake, most of the CPPs trapped into the endosomal vesicles. After escaping from endosomes, these peptides released into the cytoplasm and subsequently reach to different locations inside the cell. We have incorporated information on sub-cellular localization of CPPs. 250 peptides localized both in the cytoplasm and nucleus after the internalization, 114 peptides restricted to the cytoplasm/cytosol only, while 47 peptides localized into the nucleus. 56 peptides showed punctuated vesicular distribution. We have also incorporated information on uptake efficiency of CPPs. In literature, data on uptake efficiency of various CPPs has been presented in comparison with some positive control (i.e. Tat, penetratin, pVEC etc.). So, to make it simple, we have classified uptake efficiencies of CPPs into three categories: Low (<25% relative to control); medium (between 26% and 75% relative to control); and high (>75% relative to control).
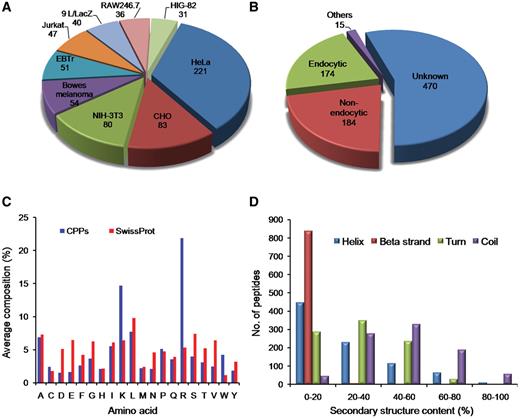
Distribution of CPPs in CPPsite. (A) distribution of peptides based on cell lines, (B) distribution of CPPs based on uptake mechanism, (C) average amino acid composition of peptides and (D) distribution of peptides based on secondary structure composition.
We have computed average amino acid composition of these peptides and observed that certain types of residues (Arg, Lys, Leu and Ala) are more abundant (Figure 3C) in CPPs. We have also computed the average amino acid composition of a set of large number of proteins extracted from SwissProt and compared with amino acid composition of CPPs. It was observed that Arg and Lys residues are more abundant in CPPs compared to SwissProt sequences. This is in agreement with various studies, which showed that positively charged residues are usually preferred in CPPs. These cationic peptides interact with negatively charged phosphates and sulfates on the surface of cell.
Since the structure of the CPPs play an important role in internalization, we have computed secondary structure of each peptide and classified peptides based on secondary structure content. As shown in Figure 3D, most of the peptides have <20% helix and β-strand content. Only limited peptides have helix or β-strand content >80%. In case of turn, it is observed that peptides having turns up to 60% are equally distributed. In case of coil, a large number of peptides have coil content in the range of 20–80%. This is expected as it is difficult for a small peptide to maintain a regular secondary structure.
Discussion
In the past few years, due to the huge therapeutic potential of CPPs, a growing interest has been seen in CPP based research. Most of the research on CPPs is currently being focused on rational design of peptide based drugs and diagnostics. All over the world, there is a healthy competition among the researchers to design novel and effective CPPs. Since the discovery of Tat, thousands of research papers have been published, illustrating the characterization and therapeutic application of various novel CPPs. This information is very useful for researchers/scientists to design novel CPPs, and further characterization of the existing CPPs. Information about CPPs is scattered in literature, it is difficult to access this useful information. CPPsite is a first comprehensive database of its kind, which provides both physicochemical and experimental information related to CPPs.
CPPsite will be useful for researchers in many ways: (i) users can check whether their peptides of interest are already reported as CPP or not; (ii) users can select the best CPP from CPPsite with the desired efficiency and physico-chemical properties for delivery; (iii) users can exploit structural information of CPP for docking or molecular dynamics of the peptide-membrane complex. In addition, CPPsite database will be helpful for developing prediction methods for CPP. In conclusion, CPPsite, the freely available open source database, would be very useful to scientific community working in the field of peptide based drug discovery.
Update of CPPsite
The web server allows users to submit new entry of CPPs on-line by filling HTML form. However, before including in CPPsite, we will confirm the validity of new entry in order to maintain the quality. Our team is also searching and adding new entries of CPPs in database from published literature. Attempts will be made to update this database regularly.
Limitations and future developments
The unique feature of CPPsite is that it provides structural information of all CPPs. We have predicted structure of peptides that contain only natural amino acids. In CPPsite, there are number of CPPs, which have non-natural or d-amino acids. Since there is no method available presently, which can predict the structure of peptides having non-natural and d-amino acids, therefore, we have predicted the structure of such peptides by removing non-natural amino acids or by converting d-amino acids to l-amino acids. One of the limitation of peptide based drug delivery or therapy is their stability or half life in vivo. In future, attempts will be made to provide half life of these CPPs.
Availability and requirements
CPPsite is available at http://crdd.osdd.net/raghava/cppsite/.
Funding
The authors would like to thank Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology (DBT), Govt. of India for the financial support. Funding for open access charge: Open Source for Drug Discovery Consortium (CSIR, India).
Conflict of interest. None declared.



