-
PDF
- Split View
-
Views
-
Cite
Cite
Sarah Burge, Elizabeth Kelly, David Lonsdale, Prudence Mutowo-Muellenet, Craig McAnulla, Alex Mitchell, Amaia Sangrador-Vegas, Siew-Yit Yong, Nicola Mulder, Sarah Hunter, Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation, Database, Volume 2012, 2012, bar068, https://doi.org/10.1093/database/bar068
Close - Share Icon Share
Abstract
InterPro amalgamates predictive protein signatures from a number of well-known partner databases into a single resource. To aid with interpretation of results, InterPro entries are manually annotated with terms from the Gene Ontology (GO). The InterPro2GO mappings are comprised of the cross-references between these two resources and are the largest source of GO annotation predictions for proteins. Here, we describe the protocol by which InterPro curators integrate GO terms into the InterPro database. We discuss the unique challenges involved in integrating specific GO terms with entries that may describe a diverse set of proteins, and we illustrate, with examples, how InterPro hierarchies reflect GO terms of increasing specificity. We describe a revised protocol for GO mapping that enables us to assign GO terms to domains based on the function of the individual domain, rather than the function of the families in which the domain is found. We also discuss how taxonomic constraints are dealt with and those cases where we are unable to add any appropriate GO terms. Expert manual annotation of InterPro entries with GO terms enables users to infer function, process or subcellular information for uncharacterized sequences based on sequence matches to predictive models.
Database URL:http://www.ebi.ac.uk/interpro. The complete InterPro2GO mappings are available at: ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/external2go/interpro2go
Introduction
The InterPro database (1) is an integrated resource of predictive protein signatures. These signatures use a range of computational methods to infer potential structure, function and/or evolutionary relationships for a query sequence. Equivalent signatures are grouped together in the same InterPro entry, and each entry contains information about the proteins matched by these signatures, including manual annotation, and links to related resources to provide enhanced biological context. Each InterPro entry is assigned a type depending on what the entry describes: family (a group of proteins with a common evolutionary origin), domain (a distinct functional, structural or sequence unit), site (which may be further subdivided into active site, binding site, conserved site or post-translational modification) and repeat (full definitions of InterPro entry types are via the user documentation available at: http://www.ebi.ac.uk/interpro/). A protein sequence may match several InterPro entries; for example, it may have matches to entries describing its N- and C-terminal domains, as well as to entries describing the protein family to which it belongs. Some InterPro entries are also organized in hierarchies, which are used to link more general entries (termed the parent entry) to more specific entries (known as child entries). The database is searchable by a range of identifiers, or by sequence using InterProScan (2). InterPro data are frequently used by genome/proteome sequencing projects to assist in characterization of putative gene products (3), and are widely included in pipelines for annotation of sequences from next-generation sequencing efforts (4).
The Gene Ontology (GO) Consortium provides a controlled vocabulary that can be used to describe gene products in a consistent and structured fashion (5). The GO is the most widely used biomedical ontology and the utility of GO annotations is highlighted by the number of resources that provide them, including major sequence databases [such as UniProtKB (6)] and many of the prominent model organism databases (7). The GO consists of three structured ontologies, describing Molecular Function, Biological Process and Cellular Component. Terms are related to each other by well-defined relationships, and are provided with stable, unique identifiers and explicit, consistent descriptions. GO terms are assigned to genes or gene product identifiers by biological database annotation efforts by manually extracting evidence from published experimental data, inferring annotations based on homology or via a range of computational inference methods. The nature of the evidence used to assign a GO term to a given protein is indicated by an evidence code.
Manual annotation of individual gene product sequences from the literature provides the gold standard of functional annotation, but it is a time-consuming approach. The rapidly increasing amount of sequence data for diverse organisms means that automated annotation plays an essential role in predicting gene product behaviour. InterPro's aim is to provide high-quality automatic annotation, based on experimental evidence. GO annotation provided by InterPro is the largest source of automatic GO annotation for proteins from all organisms, (e.g. as of UniProtKB-GOA v101, it supplies 66% of the GO annotations for UniProtKB proteins, providing over 56 million distinct annotations) and is used by many annotation communities to supplement their manual annotation work. Importantly, InterPro GO annotation allows users to infer information about an uncharacterized sequence based on match(es) of that sequence to a GO-annotated InterPro entry. This process enables transfer of information from evolutionarily related sequences that have been characterized experimentally. InterPro has been producing GO annotations since 2002, and the InterPro approach to GO annotation, its benefits and limitations are described in this article.
Methods
GO terms are assigned to the InterPro entry, not to the individual sequence
A cornerstone of the InterPro GO annotation protocol is that curators annotate an InterPro entry, and not to the individual sequence; this is the key difference between InterPro GO annotations and those provided by manual annotation efforts. GO terms are assigned by a curator to an InterPro entry based on the common characteristics of the protein set matched by the signatures belonging to that entry. InterPro2GO annotations all apply the GO evidence code ‘Inferred from Electronic Annotation’ (IEA), indicating that the GO annotations are the result of an automated prediction pipeline and have not been individually reviewed by curators. An individual sequence will therefore inherit an InterPro GO term if it matches the signatures within the InterPro entry when searched against them.
GO terms assigned to InterPro entries must apply to the majority of proteins in the entry
InterPro entries annotate all sequences that match the computational signature(s) contained in the entry; entries may contain signatures describing a small set of proteins with high-functional specificity (as in the case of IPR004025: fungal ribotoxin that matches 34 proteins), or they may contain signatures describing a large and functionally diverse family (as in the case of IPR011701: major facilitator superfamily that matches 1 08 611 proteins). It is only possible to transfer GO annotations from the UniProtKB record of a protein if those terms are considered to be applicable to all the other sequences associated with the entry. Large and diverse families may contain proteins with many annotations that are too specific to apply to the entire InterPro entry.
General protocol
A flowchart illustrating the InterPro curator protocol is presented in Figure 1. When annotating an InterPro entry, a curator first identifies those UniProtKB/Swiss-Prot (i.e. reviewed) sequences matched by the entry that has been experimentally characterized. Based on this information, the curator considers whether each of the GO terms that could potentially be applied is valid for the remaining proteins in the match set. This is done by evaluating alignments of the sequences and the experimental evidence in the literature. The UniProtKB/Swiss-Prot GO terms should be applicable to at least 95% of reviewed proteins in the entry. This cut-off sets a stringent standard for evidence yet provides enough flexibility to accommodate the predictive nature of the signatures used in creating InterPro entries. More stringent requirements would result in a loss of a large number of valid InterPro2GO mappings. InterPro GO coverage as of InterPro v34.0 is detailed in Table 1.
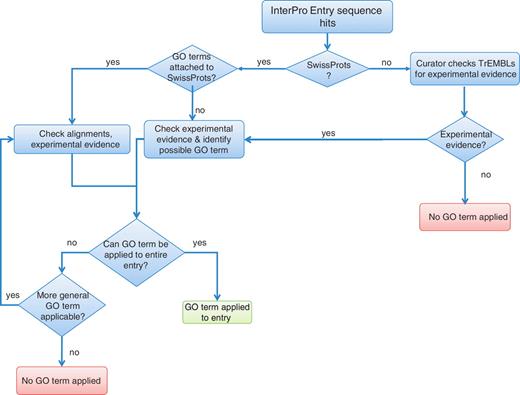
Flowchart outlining the decision process taken by InterPro curators in order to assign GO terms.
| InterPro2GO, v 34.0 . | Entries . | Coverage (%) . |
|---|---|---|
| Number of InterPro entries | 22 245 | 100 |
| Associated with at least one GO term | 10 721 | 46.2 |
| Unmapped entries | 11 974 | 54.8 |
| Of which conserved sites | 634 | 2.9 |
| Other unmappable entries | 3335 | 15.0 |
| Number of unique GO terms | 3568 | |
| Number of individual sequences annotated | 11 515 689 |
| InterPro2GO, v 34.0 . | Entries . | Coverage (%) . |
|---|---|---|
| Number of InterPro entries | 22 245 | 100 |
| Associated with at least one GO term | 10 721 | 46.2 |
| Unmapped entries | 11 974 | 54.8 |
| Of which conserved sites | 634 | 2.9 |
| Other unmappable entries | 3335 | 15.0 |
| Number of unique GO terms | 3568 | |
| Number of individual sequences annotated | 11 515 689 |
| InterPro2GO, v 34.0 . | Entries . | Coverage (%) . |
|---|---|---|
| Number of InterPro entries | 22 245 | 100 |
| Associated with at least one GO term | 10 721 | 46.2 |
| Unmapped entries | 11 974 | 54.8 |
| Of which conserved sites | 634 | 2.9 |
| Other unmappable entries | 3335 | 15.0 |
| Number of unique GO terms | 3568 | |
| Number of individual sequences annotated | 11 515 689 |
| InterPro2GO, v 34.0 . | Entries . | Coverage (%) . |
|---|---|---|
| Number of InterPro entries | 22 245 | 100 |
| Associated with at least one GO term | 10 721 | 46.2 |
| Unmapped entries | 11 974 | 54.8 |
| Of which conserved sites | 634 | 2.9 |
| Other unmappable entries | 3335 | 15.0 |
| Number of unique GO terms | 3568 | |
| Number of individual sequences annotated | 11 515 689 |
If the UniProtKB/Swiss-Prot GO terms are too specific to be attached to an entire InterPro entry, the InterPro curator can choose a related but more general GO term that is nonetheless still applicable to the full set of sequences. If no GO term exists to describe the function, creation of an appropriate term is requested from the GO consortium. If there is no experimental evidence to confirm a function, process or location term that can be applied to all sequences in the entry, then no GO term is applied.
While UniProtKB/Swiss-Prot annotations are used as a starting point, we are not limited to these terms: unreviewed proteins in UniProtKB/TrEMBL are included for consideration if there is sufficient experimental evidence in support of a particular GO term. Similarly, if a curator identifies a function, process or location in the literature, which is applicable to the entire InterPro entry protein match set but which is not currently annotated to any individual sequence by UniProtKB, the appropriate term is added to the entry. GO annotations by TIGRFAMs (8), HAMAP (9) and PANTHER keywords (10) are also considered for annotation, and are reviewed by a curator before inclusion. Once GO terms have been chosen, the InterPro abstract is updated with references to the literature supporting the annotation. With the exception of conserved sites (where there is an implicit lack of experimental evidence detailing involvement in functions, locations or processes), the above protocol currently applies to all InterPro entry types; however, some changes (detailed below) now occur for domains.
InterPro GO annotations are available to the community primarily in two forms: users may query a sequence or sequences using InterProScan, or browse and download mappings at the InterPro website. InterPro GO annotations are also available at a sequence level via UniProt-GOA.
InterPro and GO data structures are complementary
More specific family or domain entries, located at the leaf nodes of InterPro hierarchies (and which therefore might only describe a few well-characterized proteins) may be annotated with a correspondingly specific GO term. Conversely, more general InterPro family and domain entries may be annotated with a more general GO term, subject to meeting evidence requirements.
In Figure 2, we present an example of InterPro GO mapping, as applied to family entries, which illustrates the requirement for evidence and the complementary nature of the InterPro and GO data structures. The InterPro entry ‘Glycosyl transferase, family 9’ (IPR002201) is mapped to the molecular function term ‘transferase activity, transferring glycosyl groups’ (GO:0016757), while its child entry ‘Lipopolysaccharide heptosyltransferase I’ (IPR011908) is annotated with the more specific ‘Lipopolysaccharide heptosyltransferase activity’ (GO:0008920). However, another child entry of the ‘Glycosyl transferase, family 9’ represents ‘Lipopolysaccharide heptosyltransferase III, putative’ (IPR011916) and has not been assigned more specific GO annotation because although the signature does match reviewed proteins, no experimental evidence is available in the literature to support their function.
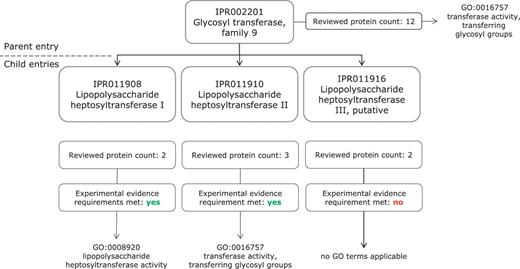
Application of GO molecular function terms to IPR002201 and its child entries. IPR002201 is a more general entry, which encompasses the proteins matched by its three child entries, IPR011908, IPR011910 and IPR011916. The increased specificity of the child entry can be reflected in the GO annotation; IPR011908 has a more specific Molecular Function term than the parent entry IPR002201.
Improved GO annotation of InterPro domain entries
Historically, InterPro entries of type domain were assigned GO terms from the protein families in which the domain was found, and not based on the function of the specific domain that the entry describes (11). This potentially could lead to the domain being incorrectly annotated with the function of another domain with which it co-occurs in a given protein family. Henceforth, GO terms will be applied to domains according to published experimental evidence of the domain's specific function. Otherwise, the curation procedure is identical to that outlined in the general protocol.
Quality control
The predictive nature of the signatures contained within InterPro means that inappropriate matches (false positives) to InterPro signatures occasionally occur. A protein that has obtained an incorrect GO annotation by virtue of a false positive match to an InterPro entry (so long as that InterPro entry is itself correctly GO annotated) will be passed on to UniProtKB-GOA (12). The InterPro GO annotation for that individual sequence may then be annotated with a NOT qualifier, and this information made available at the UniProtKB-GOA webpage for the sequence.
Additionally, some GO terms have taxonomic constraints, i.e. they may only be applied to proteins belonging to certain taxonomic groups (13). These taxonomic restrictions are a GO resource and are used in collaboration with the UniProtKB-GOA annotation project. The taxonomic constraints developed by the GO Consortium are broadly defined as two types: only_in and never_in. The only_in constraint means that a given GO term may only be applied to gene products from the specified taxonomic grouping, while the never_in constraint means that the GO term must not be applied to gene products from the specified taxonomic groups. Prior to each release, InterPro GO terms that violate these constraints are checked for. We also check automatically for redundant terms, such as cases where two GO terms with the same path to the root term have been applied to a single entry. Terms appearing in these automatic checks are referred for manual curation.
Given the sheer volume of sequence space that InterPro covers, we rely heavily on communications from our users to alert us to incorrect individual GO mappings. Users who identify incorrect mappings or wish to suggest possible GO terms may notify InterPro curators through the support channels on the InterPro website. Feedback from users who have identified GO terms that are incorrect or too specific enables constant refinement of the mappings.
p53 as a case study of InterPro GO annotation
The p53 family of tumour suppressors is well studied due to its central role in human diseases. In mammals, p53 drives the transactivation of apoptosis-inducing genes and therefore plays a key role in triggering appropriate cell death based on injury or other cell insult (14). Proteins in the p53 family consist of a DNA-binding domain and a tetramerization domain; family members also have a transactivation domain, however, there are ΔN isoforms that lack transactivation activity (15). Furthermore, in p63 and p73 family members, a large number of C-terminal splice variants exist that add considerable functional and structural diversity. In Figure 3, we have used the tumour suppressor p53 family of proteins to illustrate GO annotation within InterPro. Note that all accessions and protein counts used in this example are referring to release 34.0 of InterPro.
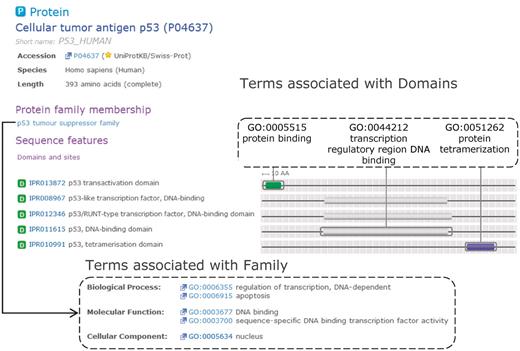
Complementary domain and family GO mapping for InterPro entries that match the human cellular tumour antigen p53. Domain GO annotation enables the function(s) of the family to be attributed to individual domains within the protein.
The most specific family entry containing the Homo sapiens p53 tumour suppressor (UniProtKB accession: P04637) is ‘p53 tumour suppressor family’ (IPR002117), containing 331 proteins. This entry covers several different isoforms of p53, p63 and p73. Due to its role as a transcriptional activator, the p53 family has GO terms attached to it that describe various aspects of this process: ‘regulation of transcription, DNA dependent’ (GO:0006355), ‘DNA binding’ (GO:0003677), ‘sequence-specific DNA binding transcription factor activity’ (GO:0003700), ‘apoptosis’ (GO:0006915) and ‘nucleus’ (GO:0005634). As the InterPro entry describes both ΔN and TA isoforms, we are unable to apply the more specific ‘positive regulation of apoptosis’ (GO:0043065) or ‘negative regulation of apoptosis’ (GO:0043066), as application of these terms would be incorrect for a significant fraction of the proteins contained in this entry, violating the previously described 95% guideline.
The three InterPro domains matching p53 provide GO annotation that is complementary to the family annotation. The p53 transactivation domain represented by IPR013872 is currently only mapped to the ‘protein binding’ (GO:0005515) term as there is currently no GO term that adequately covers the role this domain plays in binding co-activators such as p300. The p53 DNA-binding domain (IPR011615) is mapped to ‘transcription regulatory region DNA binding’ (GO:0044212). Under the new domain mapping guidelines, it would not be mapped to (for example) ‘sequence-specific DNA binding transcription factor activity’ (GO:0003700), as this behaviour is only exhibited by the whole protein, and is not solely due to this domain acting independently. Finally, the p53 C-terminal tetramerization domain (IPR010991) is mapped to ‘protein tetramerization’ (GO:0051262). By combining GO annotations from domain and family entries that a protein matches, users can identify which domains are responsible for particular elements of protein family function. This example illustrates how a domain-based approach to GO mapping leads to a more accurate and useful association of GO terms to proteins.
Summary
Increasing volumes of genomic and meta-genomic data from high-throughput sequencing technologies means that annotation of gene products remains a bottleneck, and that automated methods are increasingly important for our interpretation of this wealth of data. InterPro GO annotations provide a valuable means of annotating sequences about which little is known experimentally, based as far as possible on experimental evidence of homologous sequences. The InterPro2GO mappings produce high-quality GO annotations to individual sequences that are based on a combination of experimental evidence and sequence analysis. We aim to give InterPro's data a functional, structural and evolutionary context to ensure its continued utility to the biological community and the GO annotation process is crucial to achieving this aim.
Funding
European Union under the program ‘FP7 capacities: Scientific Data Repositories’ (grant number 213037). The project is entitled IMproving Protein Annotation and Co-ordination using Technology (IMPACT). BBSRC Bioinformatics and Biological Resources Fund (grant BB/F010508/1). Funding for open access charge: EMBL.
Conflict of interest. None declared.
Acknowledgements
We thank Emily Dimmer and Claire O'Donovan for their critical reading of this manuscript and Tony Sawford for assistance with protein match counts.



