-
PDF
- Split View
-
Views
-
Cite
Cite
Kevin L. Childs, Kranti Konganti, C. Robin Buell, The Biofuel Feedstock Genomics Resource: a web-based portal and database to enable functional genomics of plant biofuel feedstock species, Database, Volume 2012, 2012, bar061, https://doi.org/10.1093/database/bar061
Close - Share Icon Share
Abstract
Major feedstock sources for future biofuel production are likely to be high biomass producing plant species such as poplar, pine, switchgrass, sorghum and maize. One active area of research in these species is genome-enabled improvement of lignocellulosic biofuel feedstock quality and yield. To facilitate genomic-based investigations in these species, we developed the Biofuel Feedstock Genomic Resource (BFGR), a database and web-portal that provides high-quality, uniform and integrated functional annotation of gene and transcript assembly sequences from species of interest to lignocellulosic biofuel feedstock researchers. The BFGR includes sequence data from 54 species and permits researchers to view, analyze and obtain annotation at the gene, transcript, protein and genome level. Annotation of biochemical pathways permits the identification of key genes and transcripts central to the improvement of lignocellulosic properties in these species. The integrated nature of the BFGR in terms of annotation methods, orthologous/paralogous relationships and linkage to seven species with complete genome sequences allows comparative analyses for biofuel feedstock species with limited sequence resources.
Database URL:http://bfgr.plantbiology.msu.edu
Introduction
With growing interest in the utilization of plant biomass for the production of ethanol and other biofuels, the use of plant species as biofuel feedstocks has become a research focal point. However, due to concerns about diverting grain and seed from human and livestock feed to biofuel feedstock production, emphasis has shifted to the use of lignocellulose-derived biofuel production, and research is now directed at improving not only lignocellulosic yield but also quality traits in these species (1–3).
One key step in agronomic trait improvement relevant to biofuel feedstock production is identifying and understanding the genetic factors involved in the production and regulation of yield and quality traits. However, while many species have been considered for use as lignocellulosic biofuel feedstocks (4–9), only Populus trichocarpa, Sorghum bicolor and Zea mays have sequenced genomes with accompanying annotation resources that can be used to enable genome-assisted crop improvement (10–14). Currently, genome sequencing efforts are in progress for a number of other biofuel feedstock species including Miscanthus × giganteus, Panicum virgatum and Pinus taeda (http://www.jgi.doe.gov/genome-projects/; http://pinegenome.org/pinerefseq/). However, for a wide range of biofuel feedstock species, access to genic regions is limited to transcript sequence resources in the form of assembled and annotated Sanger-generated Expressed Sequence Tags (ESTs) (15–17). Although the methods used by these various genome and transcriptome annotation projects differ, they typically include sequence alignments to genes and transcript sequences from other species, protein domain identification, gene ontology (GO) assignments (18), gene family computations and functional descriptions. All of these provide an initial estimation of gene function and enable functional genomics.
Access to genome and transcriptome sequences from multiple species permits comparative analyses that are highly informative in determining gene function at either the bioinformatic or the experimental level. Comparative analyses between closely related and more distantly related species are both useful. With more closely related species, clade-specific genes can be identified, but comparative analyses involving more distantly related species permit the identification of highly conserved genes that may have roles in core biological processes. Comparative analyses are essential for species lacking a genome sequence as is the case for a large number of biofuel feedstock species, and in this report, we describe the Biofuel Feedstock Genomic Resource (BFGR), a database that provides high-quality, uniform, integrated and comparative functional annotation of gene and transcript assembly sequences from species of interest to lignocellulosic biofuel researchers. The annotated sequences include genes from seven species with sequenced genomes and transcript assemblies from an additional 47 biofuel and biofuel-related species. All sequences have been uniformly annotated and assigned functional descriptions. Annotation includes BLAST alignments (19) to UniRef proteins (20) and the proteomes of seven plant species with sequenced genomes in addition to InterPro protein domain analysis (21). Where possible, sequences have been mapped to KEGG metabolic pathways (22). Analyses have been performed to identify Simple Sequence Repeats (SSRs) from all sequences and Single Nucleotide Polymorphisms (SNPs) from the transcript assembly sequences to provide researchers with candidate genetic markers. Most importantly, ortholog analysis has been performed on all sequences so as to facilitate identification of orthologous and paralogous sequences from closely related species thereby leveraging data between species. The BFGR database also includes information about sequence resources, expression data sets, Pubmed records and germplasm resources. No other database is similarly focused on providing such a broad and fully integrated annotation of sequences from biofuel feedstock and related species.
Materials and methods
Species and sequences analyzed in BFGR
The species in the BFGR database include seven species with sequenced genomes (Arabidopsis thaliana, Brachypodium distachyon, P. trichocarpa, Oryza sativa, S. bicolor, Vitis vinifera and Z. mays) and 47 additional species that are of direct interest to lignocellulosic biofuel researchers, are related to biofuel species, serve as model or reference genomes for major taxa, or have woody growth habits (Table 1). Transcript and protein sequences for model genomes were obtained from their respective sequencing projects (12–14,23–26). For the remainder of the species, PlantGDB PUT (putative unique transcript) sequence assemblies were used as proxy gene sets (15). To remove low quality sequences, we filtered all PUT sequences to remove assemblies that were shorter than 250 nt, that had >10 N's, or that had >50% low-complexity sequence as determined by the seg low-complexity filter program. The remaining transcript assemblies were translated by ESTScan3 (27) using appropriate custom built codon usage matrices from either A. thaliana, O. sativa or a combined matrix of Pinaceae species: Piceae sitchensis, Picea glauca and P. taeda.
| Species . | Sequence source . | Sequence version . | Number filtered sequencesa . | Number with functional annotationa . | Number assigned to ortholog group . | Number assigned to KEGG ortholog . | Number unique KEGG pathways . |
|---|---|---|---|---|---|---|---|
| Agrostis stolonifera | PlantGDBb | 173a | 7732 | 3801 | 3922 | 2118 | 219 |
| Arabidopsis thaliana | TAIRc | TAIR 9 | 33 410 (27 379) | 24 698 (19 878) | 29 481 | 15 493 | 244 |
| Avena barbata | PPlantGDBb | 173a | 20 182 | 14 885 | 14 081 | 8505 | 238 |
| Avena sativa | PlantGDBb | 173a | 11 800 | 7492 | 7614 | 4387 | 230 |
| Brachypodium distachyon | Phytozomed | Bradi_1.0 | 32 255 (25 532) | 28 353 (22 110) | 29 262 | 15 598 | 247 |
| Cenchrus ciliaris | PlantGDBb | 165a | 11 688 | 7753 | 7707 | 4348 | 229 |
| Cryptomeria japonica | PlantGDBb | 167a | 23 013 | 12 167 | 11 694 | 6603 | 238 |
| Cynodon dactylon | PlantGDBb | 169a | 10 660 | 5831 | 6140 | 3289 | 228 |
| Eragrostis curvula | PlantGDBb | 165a | 8375 | 4072 | 3737 | 2657 | 213 |
| Festuca arundinacea | PlantGDBb | 175a | 30 668 | 19 074 | 16 973 | 10 810 | 242 |
| Festuca pratensis | PlantGDBb | 173a | 30 614 | 17 572 | 16 050 | 9696 | 244 |
| Helianthus annuus | PlantGDBb | 169a | 53 457 | 32 990 | 27 027 | 18 379 | 243 |
| Helianthus argophyllus | PlantGDBb | 157a | 18 597 | 13 154 | 12 009 | 7365 | 237 |
| Helianthus ciliaris | PlantGDBb | 59a | 16 090 | 12 084 | 11 909 | 6834 | 241 |
| Helianthus exilis | PlantGDBb | 157a | 21 276 | 15 072 | 14 281 | 8351 | 239 |
| Helianthus paradoxus | PlantGDBb | 159a | 19 082 | 13 088 | 12 020 | 7520 | 242 |
| Helianthus petiolaris | PlantGDBb | 157a | 13 727 | 9431 | 9158 | 5396 | 236 |
| Helianthus tuberosus | PlantGDBb | 159a | 25 371 | 18 562 | 17 304 | 10 273 | 241 |
| Hordeum vulgare | PlantGDBb | 169a | 95 386 | 60 230 | 42 347 | 32 769 | 248 |
| Leymus cinereus × Leymus triticoides | PlantGDBb | 163a | 12 712 | 9935 | 9888 | 5600 | 233 |
| Medicago sativa | PlantGDBb | 163a | 3423 | 2411 | 2387 | 1378 | 216 |
| Medicago truncatula | PlantGDBb | 169a | 48 316 | 29 507 | 23 876 | 15 319 | 250 |
| Oryza sativa | MSU RGAPe | Release 6.1 | 67 764 (57 168) | 48 508 (39 870) | 45 031 | 25 183 | 245 |
| Panicum virgatum | PlantGDBb | 169a | 87 504 | 55 055 | 46 586 | 27 676 | 250 |
| Picea engelmannii × Picea glauca | PlantGDBb | 157a | 13 755 | 7627 | 8362 | 4226 | 240 |
| Picea glauca | PlantGDBb | 175a | 47 231 | 27 084 | 22 392 | 14 884 | 246 |
| Picea sitchensis | PlantGDBb | 175a | 30 492 | 18 111 | 16 550 | 10 101 | 244 |
| Pinus contorta | PlantGDBb | 175a | 13 527 | 9752 | 10 319 | 5527 | 238 |
| Pinus pinaster | PlantGDBb | 157a | 11 375 | 7054 | 7396 | 4068 | 245 |
| Pinus taeda | PlantGDBb | 157a | 42 200 | 25 012 | 20 532 | 13 794 | 244 |
| Populus deltoides | PlantGDBb | 163a | 7600 | 5747 | 5908 | 3181 | 227 |
| Populus euphratica | PlantGDBb | 163a | 7894 | 5785 | 6016 | 3230 | 229 |
| Populus nigra | PlantGDBb | 163a | 30 991 | 20 268 | 18 567 | 9894 | 238 |
| Populus tremula | PlantGDBb | 163a | 15 134 | 8905 | 8820 | 4623 | 229 |
| Populus tremula × Populus alba | PlantGDBb | 169a | 11 224 | 7557 | 7216 | 4243 | 230 |
| Populus tremula × Populus tremuloides | PlantGDBb | 157a | 28 425 | 16 786 | 14 288 | 9239 | 237 |
| Populus tremuloides | PlantGDBb | 157a | 4940 | 3903 | 3715 | 2488 | 224 |
| Populus trichocarpa | Phytozomed | Version 2 | 45 778 (41 337) | 38 093 (34 122) | 37 430 | 21 004 | 246 |
| Populus trichocarpa × P. deltoides | PlantGDBb | 157a | 17 690 | 11 559 | 11 329 | 6240 | 239 |
| Populus trichocarpa × P. nigra | PlantGDBb | 157a | 9671 | 6433 | 6864 | 3350 | 236 |
| Populus × canadensis | PlantGDBb | 157a | 4548 | 3595 | 3801 | 2028 | 217 |
| Pseudoroegneria spicata | PlantGDBb | 169a | 9540 | 7344 | 7979 | 4092 | 234 |
| Pseudotsuga menziesii var. menziesii | PlantGDBb | 161a | 8753 | 3005 | 3272 | 1753 | 217 |
| Quercus petraea | PlantGDBb | 175a | 5928 | 4285 | 4384 | 2543 | 227 |
| Quercus Robur | PlantGDBb | 175a | 16 466 | 11 193 | 10 643 | 6008 | 240 |
| Saccharum officinarum | PlantGDBb | 157a | 125 666 | 78 578 | 58 154 | 41 761 | 243 |
| Secale cereale | PlantGDBb | 157a | 5298 | 3704 | 3892 | 2092 | 220 |
| Sorghum bicolor | Phytozomed | Sbi1.4 | 36 338 (34 496) | 30 460 | 30 156 | 15 241 | 243 |
| Sorghum propinquum | PlantGDBb | 157a | 8506 | 5787 | 6515 | 3123 | 226 |
| Triticum aestivum | PlantGDBb | 163b | 195 472 | 85 089 | 59 601 | 49 442 | 251 |
| Triticum monococcum | PlantGDBb | 157a | 5879 | 3909 | 4402 | 2278 | 223 |
| Triticum turgidum subsp. durum | PlantGDBb | 169a | 7203 | 4871 | 5186 | 2827 | 223 |
| Vitis vinifera | Genoscopef | Version 1 (12x) | 26 346 (26 346) | 20 652 (20 652) | 19 854 | 12 534 | 245 |
| Zea mays | MaizeSequence.orgg | 4a.53 | 53 764 (31 832) | 44 344 (25 291) | 43 541 | 24 447 | 243 |
| Species . | Sequence source . | Sequence version . | Number filtered sequencesa . | Number with functional annotationa . | Number assigned to ortholog group . | Number assigned to KEGG ortholog . | Number unique KEGG pathways . |
|---|---|---|---|---|---|---|---|
| Agrostis stolonifera | PlantGDBb | 173a | 7732 | 3801 | 3922 | 2118 | 219 |
| Arabidopsis thaliana | TAIRc | TAIR 9 | 33 410 (27 379) | 24 698 (19 878) | 29 481 | 15 493 | 244 |
| Avena barbata | PPlantGDBb | 173a | 20 182 | 14 885 | 14 081 | 8505 | 238 |
| Avena sativa | PlantGDBb | 173a | 11 800 | 7492 | 7614 | 4387 | 230 |
| Brachypodium distachyon | Phytozomed | Bradi_1.0 | 32 255 (25 532) | 28 353 (22 110) | 29 262 | 15 598 | 247 |
| Cenchrus ciliaris | PlantGDBb | 165a | 11 688 | 7753 | 7707 | 4348 | 229 |
| Cryptomeria japonica | PlantGDBb | 167a | 23 013 | 12 167 | 11 694 | 6603 | 238 |
| Cynodon dactylon | PlantGDBb | 169a | 10 660 | 5831 | 6140 | 3289 | 228 |
| Eragrostis curvula | PlantGDBb | 165a | 8375 | 4072 | 3737 | 2657 | 213 |
| Festuca arundinacea | PlantGDBb | 175a | 30 668 | 19 074 | 16 973 | 10 810 | 242 |
| Festuca pratensis | PlantGDBb | 173a | 30 614 | 17 572 | 16 050 | 9696 | 244 |
| Helianthus annuus | PlantGDBb | 169a | 53 457 | 32 990 | 27 027 | 18 379 | 243 |
| Helianthus argophyllus | PlantGDBb | 157a | 18 597 | 13 154 | 12 009 | 7365 | 237 |
| Helianthus ciliaris | PlantGDBb | 59a | 16 090 | 12 084 | 11 909 | 6834 | 241 |
| Helianthus exilis | PlantGDBb | 157a | 21 276 | 15 072 | 14 281 | 8351 | 239 |
| Helianthus paradoxus | PlantGDBb | 159a | 19 082 | 13 088 | 12 020 | 7520 | 242 |
| Helianthus petiolaris | PlantGDBb | 157a | 13 727 | 9431 | 9158 | 5396 | 236 |
| Helianthus tuberosus | PlantGDBb | 159a | 25 371 | 18 562 | 17 304 | 10 273 | 241 |
| Hordeum vulgare | PlantGDBb | 169a | 95 386 | 60 230 | 42 347 | 32 769 | 248 |
| Leymus cinereus × Leymus triticoides | PlantGDBb | 163a | 12 712 | 9935 | 9888 | 5600 | 233 |
| Medicago sativa | PlantGDBb | 163a | 3423 | 2411 | 2387 | 1378 | 216 |
| Medicago truncatula | PlantGDBb | 169a | 48 316 | 29 507 | 23 876 | 15 319 | 250 |
| Oryza sativa | MSU RGAPe | Release 6.1 | 67 764 (57 168) | 48 508 (39 870) | 45 031 | 25 183 | 245 |
| Panicum virgatum | PlantGDBb | 169a | 87 504 | 55 055 | 46 586 | 27 676 | 250 |
| Picea engelmannii × Picea glauca | PlantGDBb | 157a | 13 755 | 7627 | 8362 | 4226 | 240 |
| Picea glauca | PlantGDBb | 175a | 47 231 | 27 084 | 22 392 | 14 884 | 246 |
| Picea sitchensis | PlantGDBb | 175a | 30 492 | 18 111 | 16 550 | 10 101 | 244 |
| Pinus contorta | PlantGDBb | 175a | 13 527 | 9752 | 10 319 | 5527 | 238 |
| Pinus pinaster | PlantGDBb | 157a | 11 375 | 7054 | 7396 | 4068 | 245 |
| Pinus taeda | PlantGDBb | 157a | 42 200 | 25 012 | 20 532 | 13 794 | 244 |
| Populus deltoides | PlantGDBb | 163a | 7600 | 5747 | 5908 | 3181 | 227 |
| Populus euphratica | PlantGDBb | 163a | 7894 | 5785 | 6016 | 3230 | 229 |
| Populus nigra | PlantGDBb | 163a | 30 991 | 20 268 | 18 567 | 9894 | 238 |
| Populus tremula | PlantGDBb | 163a | 15 134 | 8905 | 8820 | 4623 | 229 |
| Populus tremula × Populus alba | PlantGDBb | 169a | 11 224 | 7557 | 7216 | 4243 | 230 |
| Populus tremula × Populus tremuloides | PlantGDBb | 157a | 28 425 | 16 786 | 14 288 | 9239 | 237 |
| Populus tremuloides | PlantGDBb | 157a | 4940 | 3903 | 3715 | 2488 | 224 |
| Populus trichocarpa | Phytozomed | Version 2 | 45 778 (41 337) | 38 093 (34 122) | 37 430 | 21 004 | 246 |
| Populus trichocarpa × P. deltoides | PlantGDBb | 157a | 17 690 | 11 559 | 11 329 | 6240 | 239 |
| Populus trichocarpa × P. nigra | PlantGDBb | 157a | 9671 | 6433 | 6864 | 3350 | 236 |
| Populus × canadensis | PlantGDBb | 157a | 4548 | 3595 | 3801 | 2028 | 217 |
| Pseudoroegneria spicata | PlantGDBb | 169a | 9540 | 7344 | 7979 | 4092 | 234 |
| Pseudotsuga menziesii var. menziesii | PlantGDBb | 161a | 8753 | 3005 | 3272 | 1753 | 217 |
| Quercus petraea | PlantGDBb | 175a | 5928 | 4285 | 4384 | 2543 | 227 |
| Quercus Robur | PlantGDBb | 175a | 16 466 | 11 193 | 10 643 | 6008 | 240 |
| Saccharum officinarum | PlantGDBb | 157a | 125 666 | 78 578 | 58 154 | 41 761 | 243 |
| Secale cereale | PlantGDBb | 157a | 5298 | 3704 | 3892 | 2092 | 220 |
| Sorghum bicolor | Phytozomed | Sbi1.4 | 36 338 (34 496) | 30 460 | 30 156 | 15 241 | 243 |
| Sorghum propinquum | PlantGDBb | 157a | 8506 | 5787 | 6515 | 3123 | 226 |
| Triticum aestivum | PlantGDBb | 163b | 195 472 | 85 089 | 59 601 | 49 442 | 251 |
| Triticum monococcum | PlantGDBb | 157a | 5879 | 3909 | 4402 | 2278 | 223 |
| Triticum turgidum subsp. durum | PlantGDBb | 169a | 7203 | 4871 | 5186 | 2827 | 223 |
| Vitis vinifera | Genoscopef | Version 1 (12x) | 26 346 (26 346) | 20 652 (20 652) | 19 854 | 12 534 | 245 |
| Zea mays | MaizeSequence.orgg | 4a.53 | 53 764 (31 832) | 44 344 (25 291) | 43 541 | 24 447 | 243 |
aNumbers in parentheses refer to gene loci from model genome species.
| Species . | Sequence source . | Sequence version . | Number filtered sequencesa . | Number with functional annotationa . | Number assigned to ortholog group . | Number assigned to KEGG ortholog . | Number unique KEGG pathways . |
|---|---|---|---|---|---|---|---|
| Agrostis stolonifera | PlantGDBb | 173a | 7732 | 3801 | 3922 | 2118 | 219 |
| Arabidopsis thaliana | TAIRc | TAIR 9 | 33 410 (27 379) | 24 698 (19 878) | 29 481 | 15 493 | 244 |
| Avena barbata | PPlantGDBb | 173a | 20 182 | 14 885 | 14 081 | 8505 | 238 |
| Avena sativa | PlantGDBb | 173a | 11 800 | 7492 | 7614 | 4387 | 230 |
| Brachypodium distachyon | Phytozomed | Bradi_1.0 | 32 255 (25 532) | 28 353 (22 110) | 29 262 | 15 598 | 247 |
| Cenchrus ciliaris | PlantGDBb | 165a | 11 688 | 7753 | 7707 | 4348 | 229 |
| Cryptomeria japonica | PlantGDBb | 167a | 23 013 | 12 167 | 11 694 | 6603 | 238 |
| Cynodon dactylon | PlantGDBb | 169a | 10 660 | 5831 | 6140 | 3289 | 228 |
| Eragrostis curvula | PlantGDBb | 165a | 8375 | 4072 | 3737 | 2657 | 213 |
| Festuca arundinacea | PlantGDBb | 175a | 30 668 | 19 074 | 16 973 | 10 810 | 242 |
| Festuca pratensis | PlantGDBb | 173a | 30 614 | 17 572 | 16 050 | 9696 | 244 |
| Helianthus annuus | PlantGDBb | 169a | 53 457 | 32 990 | 27 027 | 18 379 | 243 |
| Helianthus argophyllus | PlantGDBb | 157a | 18 597 | 13 154 | 12 009 | 7365 | 237 |
| Helianthus ciliaris | PlantGDBb | 59a | 16 090 | 12 084 | 11 909 | 6834 | 241 |
| Helianthus exilis | PlantGDBb | 157a | 21 276 | 15 072 | 14 281 | 8351 | 239 |
| Helianthus paradoxus | PlantGDBb | 159a | 19 082 | 13 088 | 12 020 | 7520 | 242 |
| Helianthus petiolaris | PlantGDBb | 157a | 13 727 | 9431 | 9158 | 5396 | 236 |
| Helianthus tuberosus | PlantGDBb | 159a | 25 371 | 18 562 | 17 304 | 10 273 | 241 |
| Hordeum vulgare | PlantGDBb | 169a | 95 386 | 60 230 | 42 347 | 32 769 | 248 |
| Leymus cinereus × Leymus triticoides | PlantGDBb | 163a | 12 712 | 9935 | 9888 | 5600 | 233 |
| Medicago sativa | PlantGDBb | 163a | 3423 | 2411 | 2387 | 1378 | 216 |
| Medicago truncatula | PlantGDBb | 169a | 48 316 | 29 507 | 23 876 | 15 319 | 250 |
| Oryza sativa | MSU RGAPe | Release 6.1 | 67 764 (57 168) | 48 508 (39 870) | 45 031 | 25 183 | 245 |
| Panicum virgatum | PlantGDBb | 169a | 87 504 | 55 055 | 46 586 | 27 676 | 250 |
| Picea engelmannii × Picea glauca | PlantGDBb | 157a | 13 755 | 7627 | 8362 | 4226 | 240 |
| Picea glauca | PlantGDBb | 175a | 47 231 | 27 084 | 22 392 | 14 884 | 246 |
| Picea sitchensis | PlantGDBb | 175a | 30 492 | 18 111 | 16 550 | 10 101 | 244 |
| Pinus contorta | PlantGDBb | 175a | 13 527 | 9752 | 10 319 | 5527 | 238 |
| Pinus pinaster | PlantGDBb | 157a | 11 375 | 7054 | 7396 | 4068 | 245 |
| Pinus taeda | PlantGDBb | 157a | 42 200 | 25 012 | 20 532 | 13 794 | 244 |
| Populus deltoides | PlantGDBb | 163a | 7600 | 5747 | 5908 | 3181 | 227 |
| Populus euphratica | PlantGDBb | 163a | 7894 | 5785 | 6016 | 3230 | 229 |
| Populus nigra | PlantGDBb | 163a | 30 991 | 20 268 | 18 567 | 9894 | 238 |
| Populus tremula | PlantGDBb | 163a | 15 134 | 8905 | 8820 | 4623 | 229 |
| Populus tremula × Populus alba | PlantGDBb | 169a | 11 224 | 7557 | 7216 | 4243 | 230 |
| Populus tremula × Populus tremuloides | PlantGDBb | 157a | 28 425 | 16 786 | 14 288 | 9239 | 237 |
| Populus tremuloides | PlantGDBb | 157a | 4940 | 3903 | 3715 | 2488 | 224 |
| Populus trichocarpa | Phytozomed | Version 2 | 45 778 (41 337) | 38 093 (34 122) | 37 430 | 21 004 | 246 |
| Populus trichocarpa × P. deltoides | PlantGDBb | 157a | 17 690 | 11 559 | 11 329 | 6240 | 239 |
| Populus trichocarpa × P. nigra | PlantGDBb | 157a | 9671 | 6433 | 6864 | 3350 | 236 |
| Populus × canadensis | PlantGDBb | 157a | 4548 | 3595 | 3801 | 2028 | 217 |
| Pseudoroegneria spicata | PlantGDBb | 169a | 9540 | 7344 | 7979 | 4092 | 234 |
| Pseudotsuga menziesii var. menziesii | PlantGDBb | 161a | 8753 | 3005 | 3272 | 1753 | 217 |
| Quercus petraea | PlantGDBb | 175a | 5928 | 4285 | 4384 | 2543 | 227 |
| Quercus Robur | PlantGDBb | 175a | 16 466 | 11 193 | 10 643 | 6008 | 240 |
| Saccharum officinarum | PlantGDBb | 157a | 125 666 | 78 578 | 58 154 | 41 761 | 243 |
| Secale cereale | PlantGDBb | 157a | 5298 | 3704 | 3892 | 2092 | 220 |
| Sorghum bicolor | Phytozomed | Sbi1.4 | 36 338 (34 496) | 30 460 | 30 156 | 15 241 | 243 |
| Sorghum propinquum | PlantGDBb | 157a | 8506 | 5787 | 6515 | 3123 | 226 |
| Triticum aestivum | PlantGDBb | 163b | 195 472 | 85 089 | 59 601 | 49 442 | 251 |
| Triticum monococcum | PlantGDBb | 157a | 5879 | 3909 | 4402 | 2278 | 223 |
| Triticum turgidum subsp. durum | PlantGDBb | 169a | 7203 | 4871 | 5186 | 2827 | 223 |
| Vitis vinifera | Genoscopef | Version 1 (12x) | 26 346 (26 346) | 20 652 (20 652) | 19 854 | 12 534 | 245 |
| Zea mays | MaizeSequence.orgg | 4a.53 | 53 764 (31 832) | 44 344 (25 291) | 43 541 | 24 447 | 243 |
| Species . | Sequence source . | Sequence version . | Number filtered sequencesa . | Number with functional annotationa . | Number assigned to ortholog group . | Number assigned to KEGG ortholog . | Number unique KEGG pathways . |
|---|---|---|---|---|---|---|---|
| Agrostis stolonifera | PlantGDBb | 173a | 7732 | 3801 | 3922 | 2118 | 219 |
| Arabidopsis thaliana | TAIRc | TAIR 9 | 33 410 (27 379) | 24 698 (19 878) | 29 481 | 15 493 | 244 |
| Avena barbata | PPlantGDBb | 173a | 20 182 | 14 885 | 14 081 | 8505 | 238 |
| Avena sativa | PlantGDBb | 173a | 11 800 | 7492 | 7614 | 4387 | 230 |
| Brachypodium distachyon | Phytozomed | Bradi_1.0 | 32 255 (25 532) | 28 353 (22 110) | 29 262 | 15 598 | 247 |
| Cenchrus ciliaris | PlantGDBb | 165a | 11 688 | 7753 | 7707 | 4348 | 229 |
| Cryptomeria japonica | PlantGDBb | 167a | 23 013 | 12 167 | 11 694 | 6603 | 238 |
| Cynodon dactylon | PlantGDBb | 169a | 10 660 | 5831 | 6140 | 3289 | 228 |
| Eragrostis curvula | PlantGDBb | 165a | 8375 | 4072 | 3737 | 2657 | 213 |
| Festuca arundinacea | PlantGDBb | 175a | 30 668 | 19 074 | 16 973 | 10 810 | 242 |
| Festuca pratensis | PlantGDBb | 173a | 30 614 | 17 572 | 16 050 | 9696 | 244 |
| Helianthus annuus | PlantGDBb | 169a | 53 457 | 32 990 | 27 027 | 18 379 | 243 |
| Helianthus argophyllus | PlantGDBb | 157a | 18 597 | 13 154 | 12 009 | 7365 | 237 |
| Helianthus ciliaris | PlantGDBb | 59a | 16 090 | 12 084 | 11 909 | 6834 | 241 |
| Helianthus exilis | PlantGDBb | 157a | 21 276 | 15 072 | 14 281 | 8351 | 239 |
| Helianthus paradoxus | PlantGDBb | 159a | 19 082 | 13 088 | 12 020 | 7520 | 242 |
| Helianthus petiolaris | PlantGDBb | 157a | 13 727 | 9431 | 9158 | 5396 | 236 |
| Helianthus tuberosus | PlantGDBb | 159a | 25 371 | 18 562 | 17 304 | 10 273 | 241 |
| Hordeum vulgare | PlantGDBb | 169a | 95 386 | 60 230 | 42 347 | 32 769 | 248 |
| Leymus cinereus × Leymus triticoides | PlantGDBb | 163a | 12 712 | 9935 | 9888 | 5600 | 233 |
| Medicago sativa | PlantGDBb | 163a | 3423 | 2411 | 2387 | 1378 | 216 |
| Medicago truncatula | PlantGDBb | 169a | 48 316 | 29 507 | 23 876 | 15 319 | 250 |
| Oryza sativa | MSU RGAPe | Release 6.1 | 67 764 (57 168) | 48 508 (39 870) | 45 031 | 25 183 | 245 |
| Panicum virgatum | PlantGDBb | 169a | 87 504 | 55 055 | 46 586 | 27 676 | 250 |
| Picea engelmannii × Picea glauca | PlantGDBb | 157a | 13 755 | 7627 | 8362 | 4226 | 240 |
| Picea glauca | PlantGDBb | 175a | 47 231 | 27 084 | 22 392 | 14 884 | 246 |
| Picea sitchensis | PlantGDBb | 175a | 30 492 | 18 111 | 16 550 | 10 101 | 244 |
| Pinus contorta | PlantGDBb | 175a | 13 527 | 9752 | 10 319 | 5527 | 238 |
| Pinus pinaster | PlantGDBb | 157a | 11 375 | 7054 | 7396 | 4068 | 245 |
| Pinus taeda | PlantGDBb | 157a | 42 200 | 25 012 | 20 532 | 13 794 | 244 |
| Populus deltoides | PlantGDBb | 163a | 7600 | 5747 | 5908 | 3181 | 227 |
| Populus euphratica | PlantGDBb | 163a | 7894 | 5785 | 6016 | 3230 | 229 |
| Populus nigra | PlantGDBb | 163a | 30 991 | 20 268 | 18 567 | 9894 | 238 |
| Populus tremula | PlantGDBb | 163a | 15 134 | 8905 | 8820 | 4623 | 229 |
| Populus tremula × Populus alba | PlantGDBb | 169a | 11 224 | 7557 | 7216 | 4243 | 230 |
| Populus tremula × Populus tremuloides | PlantGDBb | 157a | 28 425 | 16 786 | 14 288 | 9239 | 237 |
| Populus tremuloides | PlantGDBb | 157a | 4940 | 3903 | 3715 | 2488 | 224 |
| Populus trichocarpa | Phytozomed | Version 2 | 45 778 (41 337) | 38 093 (34 122) | 37 430 | 21 004 | 246 |
| Populus trichocarpa × P. deltoides | PlantGDBb | 157a | 17 690 | 11 559 | 11 329 | 6240 | 239 |
| Populus trichocarpa × P. nigra | PlantGDBb | 157a | 9671 | 6433 | 6864 | 3350 | 236 |
| Populus × canadensis | PlantGDBb | 157a | 4548 | 3595 | 3801 | 2028 | 217 |
| Pseudoroegneria spicata | PlantGDBb | 169a | 9540 | 7344 | 7979 | 4092 | 234 |
| Pseudotsuga menziesii var. menziesii | PlantGDBb | 161a | 8753 | 3005 | 3272 | 1753 | 217 |
| Quercus petraea | PlantGDBb | 175a | 5928 | 4285 | 4384 | 2543 | 227 |
| Quercus Robur | PlantGDBb | 175a | 16 466 | 11 193 | 10 643 | 6008 | 240 |
| Saccharum officinarum | PlantGDBb | 157a | 125 666 | 78 578 | 58 154 | 41 761 | 243 |
| Secale cereale | PlantGDBb | 157a | 5298 | 3704 | 3892 | 2092 | 220 |
| Sorghum bicolor | Phytozomed | Sbi1.4 | 36 338 (34 496) | 30 460 | 30 156 | 15 241 | 243 |
| Sorghum propinquum | PlantGDBb | 157a | 8506 | 5787 | 6515 | 3123 | 226 |
| Triticum aestivum | PlantGDBb | 163b | 195 472 | 85 089 | 59 601 | 49 442 | 251 |
| Triticum monococcum | PlantGDBb | 157a | 5879 | 3909 | 4402 | 2278 | 223 |
| Triticum turgidum subsp. durum | PlantGDBb | 169a | 7203 | 4871 | 5186 | 2827 | 223 |
| Vitis vinifera | Genoscopef | Version 1 (12x) | 26 346 (26 346) | 20 652 (20 652) | 19 854 | 12 534 | 245 |
| Zea mays | MaizeSequence.orgg | 4a.53 | 53 764 (31 832) | 44 344 (25 291) | 43 541 | 24 447 | 243 |
aNumbers in parentheses refer to gene loci from model genome species.
Species Overview (http://bfgr.plantbiology.msu.edu/species_list.shtml) and Sequence Summary (http://bfgr.plantbiology.msu.edu/cgi-bin/sequence_summary.cgi) pages contain species-specific sequence, publication, germplasm and genome project information. Because sequence and publication data change frequently, these data are automatically collected on a monthly basis by custom Perl scripts. In addition to the primary 54 BFGR species, Species Overview pages are also present for six species (Eleusine coracana, M. × giganteus, Oryza granulata, Pennisetum glaucum, Setaria italica and S. halepense) for which neither a complete genome sequence nor substantial PUT sequences existed in early 2010.
Alignment analyses
All sequences were processed through a common annotation pipeline. For PUT sequences, BLASTX alignments were separately performed against the predicted proteomes from the seven species with genome sequences. Additionally, the predicted proteomes from the seven species with genome sequences were aligned to each other with BLASTP. All sequences were aligned by BLAST to a custom UniRef protein database that contained all UniRef90 sequences and the higher plant proteins from UniRef100 (20). Only the best 15 alignments with an E-value <1e-10 were retained from each database. All sequences were also aligned by BLAST to the KEGG protein database. Best hits with E-values <1e-10 were used to assign sequences to KEGG Orthologs and their corresponding KEGG Pathways (22). All BLAST analyses were performed using WU-BLAST (19). InterProScan was used to run FPrintScan, ProfileScan, BlastProDom, HMMPfam, HMMSmart, HMMPanther, Gene3D, superfamily, coils and seg analyses on all PUT and sequenced genome protein sequences to identify protein signatures and possible functional sites (21); GO annotations were extracted from these results (28).
Molecular marker identification
SSRs were identified within all sequences using a custom Perl script. Mononucleotides with more than 10 repeats, dinucleotides with more than 6 repeats, and trinucleotides, tetranucleotides, pentanucleotides and hexanucleotides with more than 5 repeats were identified using simple regular expression matching. The Primer3 program was used to identify potential primers that may be used to amplify SSR molecular markers (29).
Putative SNPs were identified using custom Perl scripts. Briefly, PlantGDB-provided component sequence files of sequences that were used to generate PUT sequence assemblies and subsequence files of near exact matching duplicate sequences that were excluded from the assembly process were aligned to the PUT sequence assemblies using the vmatch alignment tool (http://www.vmatch.de). These alignments were used to form multiple sequence alignments representing each PUT sequence and were subsequently examined for polymorphic positions. Putative SNPs were only called at positions where coverage was >4× and at least two reads contained identical polymorphic bases.
Sequence mapping to expression platforms
Sequences from microarray platforms were mapped to gene and transcript sequences in the BFGR database to provide expression data on the genes and transcripts within the BFGR. Only platforms for which there are a notable number of publicly available expression data sets were included in this analysis, and therefore, microarray platform probe mapping was limited to 13 species (A. thaliana, Hordeum vulgare, Medicago truncatula, O. sativa, P. taeda, P euphratica, P. tremula × P. alba, P. trichocarpa, P. trichocarpa × P. deltoides, Saccharum officinarum, Triticum aestivum, V. vinifera and Z. mays). Platform probe sequences were downloaded from the NCBI Gene Expression Omnibus (17). For platforms with short probes, assignments to BFGR sequences were made if there was a complete (100%) match between the probe and BFGR sequences as determined by the vmatch alignment tool. For Affymetrix arrays, a probe set was assigned to a sequence if 8 out of 11 probes from a probe set matched the sequence at 100% identity and 100% coverage. For platforms that consisted of oligonucleotides greater than 60 bases or cDNAs, BLASTN was used to align probe sequences to BFGR genes and transcript assemblies. Probes were assigned to sequences for BLASTN alignments with >90% coverage and >95% sequence identity.
Ortholog and paralog analysis
While most components of the annotation pipeline were run on individual sequences, ortholog/paralog analyses were performed across species using OrthoMCL (30). Three separate sequence databases were created for use with OrthoMCL. These databases contained protein translations from the PUT transcript assemblies from either all monocot, dicot or gymnosperm species. Additionally, the protein sequences from the seven sequenced genome species were also included in each database. OrthoMCL was run with default settings to identify groups of orthologous/paralogous genes within the monocots plus the sequenced genome species, within the dicots plus the sequenced genome species and within the gymnosperms plus the sequenced genome species. Protein sequences from OrthoMCL ortholog groups were subjected to multiple sequence alignments using MUSCLE (31), and these multiple sequence alignments were used as input to proml (multiple sequences per ortholog group) or prodist (two sequences per ortholog group) from the PHYLIP package (http://evolution.gs.washington.edu/phylip.html). Due to the large number of analyses required, the Swofford and Rogers tree searching algorithm was used within proml. Newick formatted results were converted to the phyloxml format by the phyloxml_converter program (http://www.phylosoft.org), and a custom Perl script used the resulting phyloxml files to create PNG image files depicting ortholog trees as well as HTML image map files for use on the BFGR website.
Assignment of functional descriptions
Functional annotation descriptions were assigned to sequences based on either UniRef alignments or Pfam domain hits. The top 15 UniRef BLAST alignments with E-values <1e-10 were examined, and the functional descriptions of those UniRef proteins were examined for usable descriptions. If a usable description could not be computationally extracted from the top UniRef hits, then the best Pfam domain alignment with an E-value <1e-10 was used as the functional annotation. If a sequence had a significant hit to a UniRef protein but no usable UniRef or Pfam functional description could be extracted, the sequence was assigned a description of ‘Conserved gene of unknown function’ or ‘Conserved expressed gene of unknown function’ for genes and transcripts, respectively. If a sequence had no hits to either UniRef proteins or Pfam domains, then sequences were assigned functional annotations of ‘Gene of unknown function’ or ‘Expressed gene of unknown function’ for genes and transcripts, respectively. Genes from A. thaliana, O. sativa and S. bicolor were not included in this process as functional descriptions were available from the genome projects for these species.
BFGR database organization and web interface
Results from all analyses are stored in a suite of databases. For each species, a separate PostgreSQL database stores all basic sequence and annotation data. These databases use a modified chado schema (Supplementary Figure S1; refs 32,33) that contains custom tables to permit efficient retrieval of BLAST alignment results, InterPro protein domain analyses and SNP and SSR marker data. An SQLite database with a custom schema is used to store and efficiently retrieve PNG and HTML image map files that depict all gene ortholog trees and associated information as well as the data displayed on Species Overview pages (Supplementary Figure S2). An additional PostgreSQL database with a custom schema is used to provide support to all text-based searches for the BFGR website (Supplementary Figure S3). This search database makes use of text tokenization and indexing in order to quickly provide results in response to user queries.
Gbrowse, the generic genome browser (version 1.70) (34), was used to create genome browsers for each of the seven sequenced genomes. GFF files representing the gene loci and models for the genome species were obtained from the respective genome projects, and when necessary, these files were converted to GFF3 format. For each species-specific genome browser, sequences from all other BFGR species were aligned to the target species’ pseudomolecules using gap2 (35), and custom scripts parsed the gap2 alignments to GFF3 format. All genome browsers use a MySQL backend database.
The BFGR website (http://bfgr.plantbiology.msu.edu) was created to provide public access to these annotations. The website contains an overview of the project, news and FAQ pages. There are Species Overview and Sequence Summary pages as well as genome browsers for the sequenced genome species. Search tools (http://bfgr.plantbiology.msu.edu/integrated_searches.shtml) allow users to query the database based on key words, sequence identifiers, protein domain names and identifiers, GO terms, KEGG pathways, KEGG ortholog identifiers, Enzyme Commission terms, SSR characteristics and, for PUT sequences, predicted SNPs. All queries may be performed relative to individual species or across the entire suite of databases. BLAST searches are also allowed against transcript and protein sequences as well as the pseudomolecules from each of the seven sequenced genome species. The website also has a download section (ftp://ftp.plantbiology.msu.edu/pub/data/BFGR/) where users can obtain all transcript and protein sequences annotated within the BFGR. Files with functional annotation descriptions and GO term assignments can also be downloaded for each species.
The BFGR website is hosted by an Apache web server and is supported by Postgres and MySQL relational database management systems. Each of these services are hosted on separate compute servers. BLAST queries are run on an additional server. All of these servers support multiple genome resource websites. All user queries are processed by Perl CGI scripts that execute database queries, parse database outputs and deliver results as HTML pages.
Results and discussion
The major goal for BFGR was to develop a resource that not only provides high-quality, uniform and integrated annotation across sequences from multiple species but that would also permit biofuel researchers to easily perform comparative analyses on sequences from different species. The BFGR provides annotation for 1 550 736 gene and transcript assembly sequences from 54 species that include lignocellulosic biofuel feedstock species, closely related species and species with sequenced genomes (Table 1). Several species for which there is relatively little sequence data are represented by only a few thousand transcript assemblies, but for a few species, the number of transcript assemblies in the database is very large and may represent a near complete representation of the transcriptome from those species.
BFGR website
The project website (http://bfgr.plantbiology.msu.edu) contains all sequence annotations as well as pages with information about sequence, germplasm and publication data available for each BFGR species. As many investigators are comfortable viewing gene annotations through graphical genome browsers, genome browsers for seven species have been prepared that display gap2 alignments of BFGR sequences to those genomes. The primary resource at BFGR is the annotation report page that is available for each sequence within the database. Each annotation report page contains information about the gene or transcript assembly sequence, protein translation, BLAST alignments to proteins from UniRef and seven plant genomes, protein domain alignments, matches to KEGG orthologs with links to KEGG pathways, gene orthologous group results, SSR markers, predicted SNPs for PUT sequence assemblies and microarray probe matches. The annotation report pages are an important component of the BFGR not only because of their content but also because of their central position in the organization of the BFGR website (Figure 1). The results from all search pages include links to the annotation report pages of BFGR annotated sequences. Additionally, the annotation report page links to other homologous BFGR sequences via the BLAST alignment results and the orthologous gene tree results, and if a gene or PUT sequence has been mapped to a KEGG pathway, the annotation report page links to a KEGG pathway graphic with links to other sequences from that species mapped to the same KEGG pathway. Each annotation report page also contains a link to the original annotation resource.
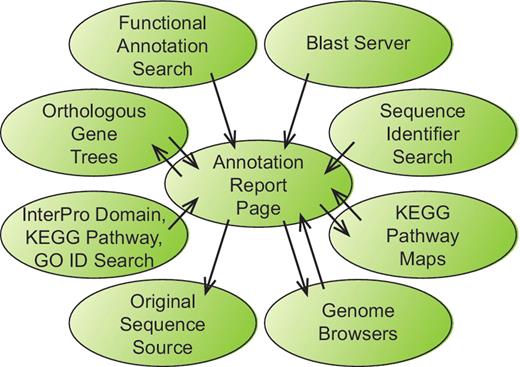
Information flow within the BFGR website. The Annotation Report Page maintains a central position within the organization of the BFGR database website. Features in tracks on the genome browsers, BLAST results and multiple other search results all provide links to sequence Annotation Report Pages. Additionally, Annotation Report Pages present graphical KEGG pathway maps and orthologous gene trees have links to the Annotation Report Pages for other sequences. Links also exist from Annotation Report Pages to the BFGR genome browsers and the original sequence source of the sequence.
Sequence mapping to KEGG pathways
A total of 545 808 BFGR annotated sequences have been mapped to KEGG orthologs, and 338 031 of these are also associated with at least one of 265 KEGG pathways (Table 1). The number of sequences within a species that map to KEGG orthologs varies roughly proportionally to the number of sequences from each species, but all species have sequences that are mapped to more than 200 different KEGG pathways. An example of the utility of the KEGG annotations within the BFGR can be seen by examining the Starch and Sucrose Metabolism pathway which utilizes 71 enzymatic processes. The number of sequences with significant homology to KEGG orthologs assigned to this pathway varies from 32 Pseudotsuga manziesii var. menziesii transcript assemblies assigned to 17 different enzymatic steps to 1405 T. aestivum transcript assemblies assigned to 35 different enzymatic steps (Supplementary Table S1). Besides being useful as a means to suggest a biochemical pathway within which a gene may function, by examining a relevant pathway, a researcher can quickly jumpstart a search for gene sequences related to a biochemistry of interest. Reviewing a pathway of interest for a particular species can also show the quality of sequence coverage within that pathway (Table 1 and Supplementary Table S1). Additionally, by examining pathways from related species, a researcher may discern whether it is likely that there are additional relevant genes that remain to be discovered in a target species.
Molecular marker analysis
Genes from sequenced genome species were analyzed for SSRs, and PUT sequence assemblies were examined for both SNPs and SSRs. PUT sequences contained 354 099 putative SNPs from 80 734 sequences (Supplementary Table S2). The numbers of each allele identified during SNP discovery are provided with SNP results so that a user may evaluate the quality of the putative SNP. SSRs were slightly less numerous with 336 653 SSRs found in 261 541 sequences (Supplementary Table S2). The base motif for each SSR and the number of times that the motif is repeated in the given sequence are given with the SSR results. If a mapping population exists, pre-computed data about SSRs and predicted SNPs can be used to develop molecular markers for mapping genes of interest.
Orthologous gene groups
While many components of the BFGR annotation pipeline are commonly used by other annotation databases, orthologous group analysis is rarely furnished, but it provides a key feature in that it not only integrates sequence analysis within BFGR but also helps to leverage existing sequence and functional knowledge across species. Ortholog analysis was separately performed using the seven genome species plus all transcript assembly predicted translations from either monocot, dicot or gymnosperm species. More than half of all BFGR sequences (887 568) were assigned to 153 229 orthologous groups (Table 1). Figure 2 provides a summary of the orthologous group results from the monocot and dicot analyses relative to the model genome species. These plots show that the majority of ortholog groups contain a large number of species and that there are hundreds of ortholog groups that contain protein sequences from almost every species used for ortholog analysis. There are fewer groups with sequences from many species as only a few of the non-model genome species have complete transcriptomes. Brachypodium distachyon tended to belong to fewer small ortholog groups, suggesting that either the B. distachyon genome is incomplete or that orthologous sequences from its most closely related species are not well represented in BFGR (Figure 2A). The large number of S. bicolor and Z. mays ortholog groups with a single additional monocot species (Figure 2A) may be due to rare gene sequences that are only represented in S. bicolor and Z. mays, the two most closely related sequenced genome species in BFGR. Populus trichocarpa tended to have sequences that are members of smaller dicot ortholog groups, and this is likely due to the large number of Populus species (11) in the database and probably reflects genus-specific sequences (Figure 2B).
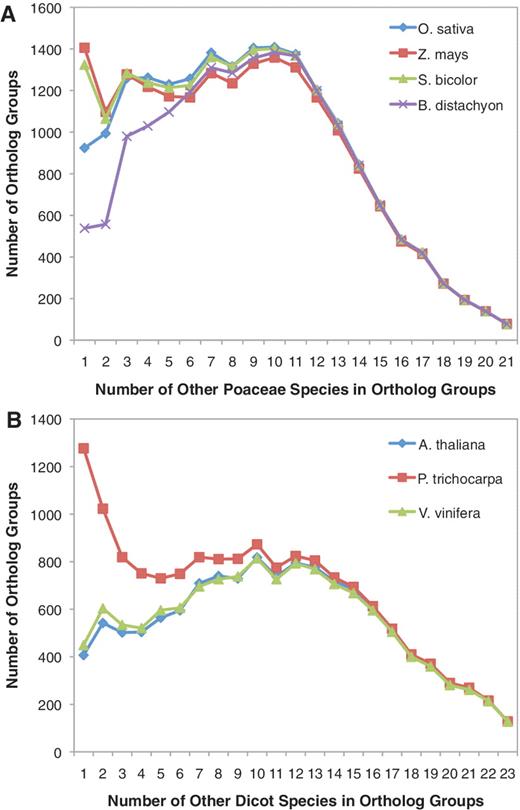
Number of additional species in orthologous groups with B. distachyon, O. sativa, S. bicolor and Z. mays. (A) For each orthologous group, the number of other Poaceae species present within the orthologous group was quantified. Counts were determined separately for orthologous groups containing B. distachyon, O. sativa, S. bicolor and Z. mays sequences. Proteins from A. thaliana, P. trichocarpa and V. vinifera were not included in these species counts. (B) For each orthologous group, the number of other dicot species present within the orthologous group was quantified. Counts were determined separately for orthologous groups containing A. thaliana, P. trichocarpa and V. vinifera sequences. Proteins from B. distachyon, O. sativa, S. bicolor and Z. mays were not included in these species counts.
Due to the large number of orthologous groups and time constraints, it was necessary to use the time-efficient Swofford and Rogers tree searching algorithm when generating the ortholog trees. Nonetheless, when examining orthologous groups of well-studied gene families, the relationships depicted in these BFGR ortholog trees are consistent with expectations. For example, the monocot orthologous gene tree for the phytochrome gene family shows three main subtrees that correspond to phytochromes A, B and C (Supplementary Figure S4). Several species have multiple gene/transcript identifiers within the three main subtrees indicating the presence of either multiple alternative transcript isoforms or distinct close paralogous genes.
Ortholog analysis in the PAL gene family
To characterize a gene family more directly relevant to lignocellulose biofuel production, members of the monocot phenylalanine ammonia-lyase (PAL) gene family were chosen for closer analysis. PAL enzymes convert phenylalanine to cinnamic acid, the first committed step in lignin production (36). Supplementary Table S3 shows the 617 monocot sequences within the BFGR database that have a functional annotation of ‘phenylalanine ammonia-lyase’ and the orthologous groups to which each belongs. The 27 PAL genes from A. thaliana, P. trichocarpa and V. vinifera are also included in Supplementary Table S3 as they were part of the OrthoMCL analysis of monocot sequences. The largest orthologous group (cluster 59) contained 127 PAL sequences. No other ortholog group had more than 13 members, but there were 396 PAL-annotated sequences that had not been assigned to any group. The average length of the proteins in cluster 59 (517 ± 197 amino acids) is notably longer than the average length of the proteins from all other clusters (220 ± 135 amino acids) and from the unassigned sequences (196 ± 87 amino acids). This suggests that incomplete gene or transcript assemblies produced truncated protein predictions were more likely to result in BLAST alignment scores that were insufficiently strong for OrthoMCL to cluster the truncated sequences with full-length members of the same gene family. There are also a few exceptions where presumably full-length protein sequences from the model genomes were either not assigned to any ortholog group or assigned to a minor cluster, and these sequences may represent PAL genes that have significantly diverged from the core PAL gene family. Given the variable completeness of gene and transcript assembly sequences in the BFGR database, the ortholog analyses are unlikely to have captured a complete picture of the ortholgous relationships in this complex gene family. This is an inherent result of working with incomplete sequence data. Nonetheless, the ortholog tree for the sequences from cluster 59 suggests interesting orthologous, paralogous and homologous relationships in this large gene family (Supplementary Figure S5). The dicot PALs are found in a single subtree, unlike the phytochrome ortholog tree, suggesting that monocot and dicot PALs have significantly diverged from each other.
Summary
The BFGR database was designed to provide highly integrative sequence annotation for not only genome but also transcriptome sequences from lignocellulosic biofuel feedstock and related species. The ease of use of the database website is an equally important feature of this resource. The user experience was an important consideration during all design and implementation decisions. Pages load quickly, and search queries have been optimized to provide fast responses. These usability features enhance the ability of researchers to readily explore the annotations of not just their target sequence but also related sequences.
Funding
United States Department of Energy (DE-FG02-08ER64631 to C.R.B. and to K.L.C.); United States Department of Agriculture (2008-04232 to C.R.B. and K.L.C.); Michigan State University AgBioResearch (to C.R.B.). Funding for open access charge: United States Department of Energy (DE-FG02-08ER64631).
Conflict of interest. None declared.
Acknowledgements
We would like to thank Brieanne Vaillancourt for assistance in preparing figures. We thank John Hamilton for providing systems administration support.



