-
PDF
- Split View
-
Views
-
Cite
Cite
Aurélie Lardenois, Alexandre Gattiker, Olivier Collin, Frédéric Chalmel, Michael Primig, GermOnline 4.0 is a genomics gateway for germline development, meiosis and the mitotic cell cycle, Database, Volume 2010, 2010, baq030, https://doi.org/10.1093/database/baq030
Close - Share Icon Share
Abstract
GermOnline 4.0 is a cross-species database portal focusing on high-throughput expression data relevant for germline development, the meiotic cell cycle and mitosis in healthy versus malignant cells. It is thus a source of information for life scientists as well as clinicians who are interested in gene expression and regulatory networks. The GermOnline gateway provides unlimited access to information produced with high-density oligonucleotide microarrays (3′-UTR GeneChips), genome-wide protein–DNA binding assays and protein–protein interaction studies in the context of Ensembl genome annotation. Samples used to produce high-throughput expression data and to carry out genome-wide in vivo DNA binding assays are annotated via the MIAME-compliant Multiomics Information Management and Annotation System (MIMAS 3.0). Furthermore, the Saccharomyces Genomics Viewer (SGV) was developed and integrated into the gateway. SGV is a visualization tool that outputs genome annotation and DNA-strand specific expression data produced with high-density oligonucleotide tiling microarrays (Sc_tlg GeneChips) which cover the complete budding yeast genome on both DNA strands. It facilitates the interpretation of expression levels and transcript structures determined for various cell types cultured under different growth and differentiation conditions.
Database URL:www.germonline.org/
Background
Microarrays have been employed to measure RNA concentrations and protein–DNA interactions for approximately 15 years (1–3) and the public certified repositories ArrayExpress (EBI) (4), Gene Expression Omnibus (NCBI) (5), and CIBEX (6) annotate, archive and disseminate these high-throughput data sets. Such repositories are crucial for data storage and meta-analyses where the output of high-throughput experiments is used for different purposes than those motivating their initial production. Recently, solutions such as the Gene Expression Atlas (7) or Genevestigator (8) have been developed which provide online access to expression data for specific genes across numerous experiments covering many species. These sources, while extremely useful and technically sophisticated, do not provide easily accessible sample annotation data important for a deeper understanding of experimental details that matter in a field. Moreover, using them to compare results from orthologous genes across species is a tedious undertaking and they cannot display information on transcript levels and structures produced with high-density oligonucleotide tiling microarrays which contain overlapping probes covering the entire genome of a given species (9,10).
To help facilitate the authoritative description of genes in the context of high-density oligonucleotide microarray (GeneChip) expression data relevant for sexual reproduction and the mitotic cell cycle across different species, we have initially developed an elaborate submission and curation system for community annotation (11–13). This system was subsequently replaced by a simpler approach based on MediaWiki, the database system underlying Wikipedia which is the most popular global knowledgebase (14,15).
Here we describe GermOnline 4.0, a gateway for manually curated high-throughput data relevant for germline development as well as the meiotic and mitotic cell cycles. The source of information reported here is of interest for basic and applied researchers in the field of developmental biology and reproduction and for biomedical researchers and clinicians working on infertility and malign tumors. We also address issues pertinent for the field of bioinformatics that focuses on high-throughput data processing and dissemination and knowledgebase development. Based on our experience over the past few years we have decided not to solicit any further contributions to GermOnlineWiki because its concept was not embraced by the community. The current release is therefore entirely dedicated to the display of high-quality expression data, protein-DNA binding data and protein–protein interaction data which have been selected on the basis of their relevance for the biomedical subjects covered by the GermOnline portal. Samples are described in detail using a recent release of our Multiomics Information Management and Annotation System (MIMAS 3.0) (16). Importantly, GermOnline 4.0 also includes the novel Saccharomyces Genomics Viewer (SGV), an online tool for interpreting complex tiling array data sets that yield genome-wide information on budding yeast transcript levels and RNA structure across many different experimental conditions.
Construction and content
Table 1 summarizes the different types of information and data available for the species currently represented in the database. We plan to include more relevant data as they are published. GermOnline 3.0 and higher were constructed using the Application Programming Interface (API) from the Ensembl genome annotation project and base web code release 50 (17). The MySQL database model is based on Ensembl and MIMAS (16). Genome sequences and annotation data were imported from Ensembl release 50 (www.ensembl.org; Homo sapiens, Pan troglodytes, Macaca mulatta, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans), TAIR release 8 (www.arabidopsis.org; Arabidopsis thaliana) and GenBank S. pombe Sanger build 1 (www.ncbi.nlm.nih.gov/genbank; Schizosaccharomyces pombe). For GermOnline 4.0 data on protein–protein interactions and genetic interactions were retrieved from BioGRID (18) and IntAct (19). Phylogenetic information on putative orthologs was assembled from Ensembl and the Orthologous Matrix project (OMA, www.cbrg.ethz.ch/oma) (20–22); information from these sources is automatically updated once a week. High-density oligonucleotide microarray probe mapping information was provided by Ensembl except in the case of Saccharomyces cerevisiae and A. thaliana where high gene density causes erroneous mapping output in Ensembl. In these cases array probes were mapped based on their sequence using the Exonerate program (23).
| . | Genome Annotation . | Gene Orthology . | 3′-UTR GeneChips . | Protein-protein interaction . | Tiling arrays . | Protein-DNA interaction . |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | * | * | * | * | * | * |
| Schizosaccharomyces pombe | * | * | * | |||
| Arabidopsis thaliana | * | * | * | * | ||
| Caenorhabditis elegans | * | * | * | |||
| Drosophila melanogaster | * | * | * | * | ||
| Danio rerio | * | * | * | |||
| Rattus norvegicus | * | * | * | * | ||
| Mus musculus | * | * | * | * | ||
| Macaca mulatta | * | * | ||||
| Pan troglodytes | * | * | ||||
| Homo sapiens | * | * | * | * |
| . | Genome Annotation . | Gene Orthology . | 3′-UTR GeneChips . | Protein-protein interaction . | Tiling arrays . | Protein-DNA interaction . |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | * | * | * | * | * | * |
| Schizosaccharomyces pombe | * | * | * | |||
| Arabidopsis thaliana | * | * | * | * | ||
| Caenorhabditis elegans | * | * | * | |||
| Drosophila melanogaster | * | * | * | * | ||
| Danio rerio | * | * | * | |||
| Rattus norvegicus | * | * | * | * | ||
| Mus musculus | * | * | * | * | ||
| Macaca mulatta | * | * | ||||
| Pan troglodytes | * | * | ||||
| Homo sapiens | * | * | * | * |
Different types of data available for species are indicated by an asterisk
| . | Genome Annotation . | Gene Orthology . | 3′-UTR GeneChips . | Protein-protein interaction . | Tiling arrays . | Protein-DNA interaction . |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | * | * | * | * | * | * |
| Schizosaccharomyces pombe | * | * | * | |||
| Arabidopsis thaliana | * | * | * | * | ||
| Caenorhabditis elegans | * | * | * | |||
| Drosophila melanogaster | * | * | * | * | ||
| Danio rerio | * | * | * | |||
| Rattus norvegicus | * | * | * | * | ||
| Mus musculus | * | * | * | * | ||
| Macaca mulatta | * | * | ||||
| Pan troglodytes | * | * | ||||
| Homo sapiens | * | * | * | * |
| . | Genome Annotation . | Gene Orthology . | 3′-UTR GeneChips . | Protein-protein interaction . | Tiling arrays . | Protein-DNA interaction . |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | * | * | * | * | * | * |
| Schizosaccharomyces pombe | * | * | * | |||
| Arabidopsis thaliana | * | * | * | * | ||
| Caenorhabditis elegans | * | * | * | |||
| Drosophila melanogaster | * | * | * | * | ||
| Danio rerio | * | * | * | |||
| Rattus norvegicus | * | * | * | * | ||
| Mus musculus | * | * | * | * | ||
| Macaca mulatta | * | * | ||||
| Pan troglodytes | * | * | ||||
| Homo sapiens | * | * | * | * |
Different types of data available for species are indicated by an asterisk
The graphical user interface and basic procedures required for database navigation and data retrieval have been described in an earlier publication (14). Minor changes include a simplified welcome page that consists of a menu to select a species (those for which microarray data are available are given in bold) and a search field for the locus query. Advanced searches for groups of genes defined in various expression profiling or functional genomics studies are possible via an implementation of EBI’s BioMart (24). A downloadable user guide and detailed help files (including tutorial videos) are also available. In addition, we regularly update ‘about’ and ‘news’ pages to help users work with the database and to alert them to new or up-coming entries.
The GermOnline report page is comprised of two sections. The ‘gene’ section covers basic information and useful links for the gene (annotation and chromosomal localization), its promoter (genome-wide in vivo DNA binding assay), phylogenetic information (including direct links to orthologs of species covered by the database) and protein interaction data. The ‘transcript’ section includes high-density oligonucleotide microarray (GeneChip) expression data, Gene Ontology annotation and information on protein domains.
Genome annotation and ortholog prediction
GermOnline 4.0 includes species for which sequenced and annotated genomes are available via the Ensembl project which provides information on the localization and the structure but not the expression of protein-coding genes (17). Model organisms were specifically selected according to their importance for researchers in the field of sexual reproduction—with emphasis on human pathologies including male and female infertility—and somatic cancer. Release 4.0 includes H. sapiens, two non-human primates (P. troglodytes, M. mulatta), two rodents (M. musculus, R. norvegicus), a fish (D. rerio), two invertebrates (C. elegans, D. melanogaster), a plant (A. thaliana) and two fungi (S. pombe, S. cerevisiae). A useful feature within the report pages (‘Orthologue prediction’) enables users to access corresponding expression data from different species represented in the database via ortholog links. We have used sequence conservation data to link report pages with each other such that users can move from one locus (and its expression data) to its (most likely) ortholog within a single mouse click. This function is critical for scientists who wish to investigate the extent to which conserved protein-coding genes show similar expression patterns in the germline and various somatic tissues across species (25).
3′-UTR GeneChip expression data
Please note that GermOnline is not an array data repository. Data are provided for viewing across experiments on a gene by gene basis. We do, however, provide a direct link to raw data available via the EBI’s ArrayExpress and the NCBI’s GEO whenever possible. We regularly include novel data sets which are either provided by members of the community or which are downloaded from the international repositories. Recent additions include a study of the human mitotic cell cycle in normal and malign cells carried out with U133 A GeneChips containing probes for all currently known human protein-coding genes (26).
The Saccharomyces Genomics Viewer
Budding yeast is the first organism for which complex data sets covering different cells types and growth and differentiation conditions are available. Sc_tlg tiling arrays provide information not only on RNA expression levels but also on transcription start- and stop sites, exon composition and antisense transcripts (9,10,27). We found the approach used for processing 3′-UTR GeneChip data to be unsuitable for displaying the output of tiling microarrays. Therefore, we developed a novel solution, the Saccharomyces Genomics Viewer (SGV), which enables users to interpret genome-wide and DNA strand-specific expression data produced with high-density oligonucleotide (Sc_tlg) tiling microarrays. Information from three different projects (10,27,28) is available for viewing online and data for individual queries are downloadable as pre-computed images in pdf format.
Users can select different query terms including the standard- and systematic-gene name, the Saccharomyces Genome Database (SGD) identifier, names of non-coding transcripts [Cryptic Unstable Transcripts (CUTs), Stable Unannotated Transcripts (SUTs) and Meiotic Unannotated Transcripts (MUTs)] chromosomal or transcript (segment) co-ordinates, or the identification numbers of autonomously replicating sequence (ARS) elements. The report page shows images covering a genomic region of 10 000 base pairs. Expression data are represented by a false-color heatmap at the single oligonucleotide probe level for samples from diploid MATa/α cells undergoing fermentation (YPD medium), respiration (YPA), sporulation (SPII 1-12 h), diploid MATα/α cells undergoing starvation (SPII 6, 10 h) as well as synchronized haploid MATa cells progressing through mitosis (27) and fermenting MATaRRP6 wild-type versus rrp6 mutant cells (10). The example shown in Figure 1a illustrates the complexity of the budding yeast transcriptome during mitotic growth and meiotic development and how high-resolution profiling puts previous results into a different perspective: GAL10 appeared to be induced during meiotic development when examined using a classical molecular biological method (29) and Ye6100 GeneChips containing probes directed against the 3′-region of the locus appeared to confirm meiotic induction (Figure 1b and c) (30). What turns out to be the case, however, is that an internal bi-directional meiotic promoter within the GAL10 locus mediates expression of divergent ncRNAs (SUT013 and a transcript that was not annotated because it overlaps with GAL10 on the same DNA strand) that are highly induced during sporulation. The conclusion is that no functional mRNA and hence Gal10 protein are present in meiotic cells which is consistent with the finding that a gal10 mutant shows no sporulation phenotype (29).
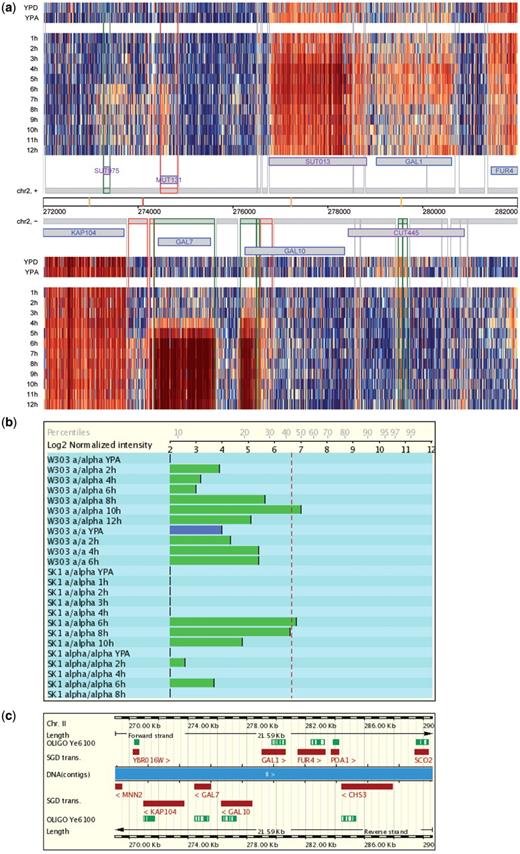
The Saccharomyces Genomics Viewer. (a) A heatmap summarizes the data for diploid MATa/α cells cultured in rich media (YPD and YPA) and sporulation medium (1–12 h). Data for the top (+) and bottom (−) strand of a region covering 10 000 bp including the GAL locus are shown as log2-transformed signals (blue and red indicating low and high values, respectively; the database report page contains a scale). Each column corresponds to an oligonucleotide probe on the tiling arrays and each line corresponds to a sample as indicated. Chromosomal co-ordinates are given. Protein coding genes are displayed in blue, and ncRNAs in purple. Transcripts are symbolized by grey boxes. Differentially expressed segments are given in green and meiosis-specific segments are shown in red. Vertical lines represent transcript boundaries. Vertical yellow and red bars represent known and predicted Middle Sporulation Element (MSE) transcription factor target motifs, respectively. (b) Graphical display of Ye6100 GeneChip expression data obtained for CDC10 in two diploid MATa/α wild-type strains (W303 and SK1) cultured in YPA and sporulation medium (W303 2, 4, 6, 8, 10, 12 h; SK1 1, 2, 3, 4, 6, 8, 10 h) as compared to sporulation-deficient controls (W303 MATa/a and SK1 MATα/α) cultured in YPA and sporulation medium (W303 2, 4, 6 h; SK1 2, 4, 6, 8 h). YPA samples are given in blue. The dotted red line represents the empirical expression-cutoff as published in (30). Log2-transformed signals and percentiles are indicated on the x-axis. (c) The GAL10 locus is shown in its genomic context and the target sequence covered by the oligonucleotide probe set present on Ye6100 is given in green (OLIGO Ye6100).
The expression data are shown in the context of S. cerevisiae’s genome annotation as of April 2009 (saccharomyces_cerevisiae.gff file from SGD). Users can continuously inspect the genome-wide data by clicking on arrows which point to the neighbouring up- or downstream regions, respectively. High resolution images from each data set are available for downloading.
Genome-wide protein-DNA binding data
Release 4.0 of the database provides Chromatin Immunoprecipitation on Chip (ChIP-Chip) data relevant for gametogenesis. This is important because the ChIP-Chip technique reveals genome-wide protein–DNA interaction patterns and therefore ideally complements expression data, especially in the case of profiling studies that compare wild-type and transcription factor mutant cells. A combination of mRNA profiling and in vivo DNA binding thus reveals direct target genes of known of potential transcriptional regulators. Release 4.0 includes a study that identified the yeast target promoters of Abf1, a general DNA binding regulator required for mitotic and meiotic functions (31) as well as work on the Sum1 repressor and the Ndt80 activator that control the expression of middle meiotic genes during yeast vegetative growth and meiotic development (32). Furthermore, we included a graphical display of selected transcription factors which are relevant for meiosis and gametogenesis from a large-scale study analyzing in vivo binding profiles of transcription factors (33). Note that the data were obtained with microarrays containing spotted PCR fragments which cover all known yeast intergenic regions and open reading frames. Data are displayed on a log2 scale with a full reference and a link to the data repository and the corresponding PubMed entry. To facilitate the interpretation of in vivo binding data that are shown as fold-enrichment of a bound DNA fragment, the corresponding data obtained with upstream and downstream fragments are also displayed; complementary expression data are available via the microarray data section in the report page. The example shown in Figure 2 for CDC3 (encoding a bud neck filament, part a) illustrates the value of the combined display of DNA binding data (parts b and c) and expression data (part d): Abf1 is required for normal expression of CDC3, it binds to the CDC3 upstream region in vivo which explains why a strain lacking fully functional Abf1 shows the same defect in cytokinesis and budding as cdc3 mutant cells [for more details, see ref. (31)].
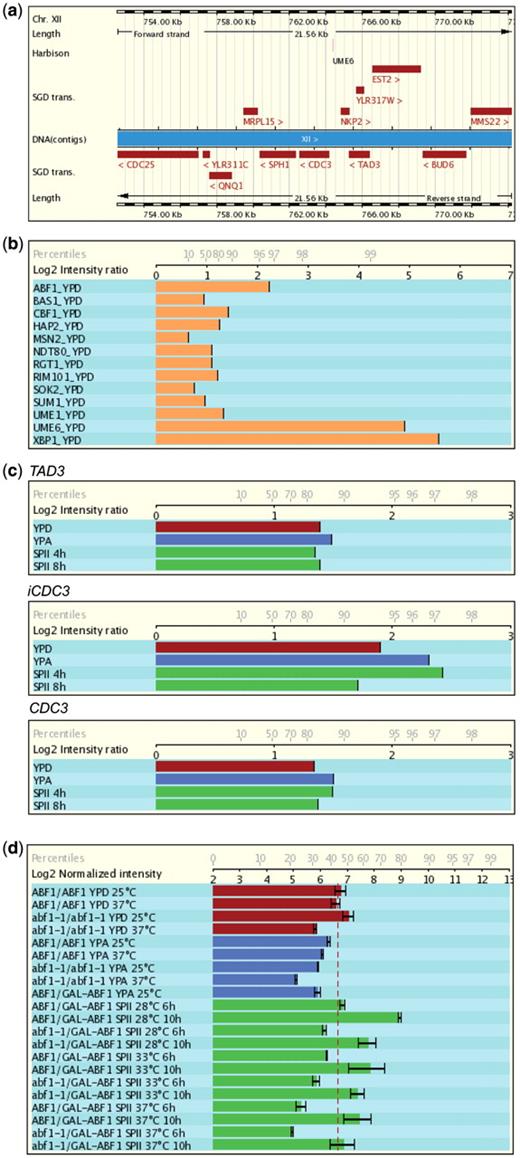
Display of genome annotation, ChIP-Chip and microarray expression data. (a) Ensembl genome annotation for CDC3 and neighbouring loci. Protein-coding genes are red rectangles. Chromosomal coordinates are given. Regulatory motifs investigated by Harbison et al. (33) are shown. (b) In vivo binding of selected transcription factors including Abf1 to the CDC3 (YLR314C) up-stream region in haploid MATa cells cultured in rich medium with glucose (YPD). (c) Abf1 binding in samples from fermenting (YPD, red), respiring (YPA, blue) and sporulating (SPII 4 h and 8 h, green) cells. Signals for the upstream gene (TAD3), the CDC3 promoter (iCDC3) and the CDC3 coding region are given for comparison. (d) CDC3 mRNA expression data in strains containing homozygous wild-type (ABF1) or temperature sensitive (ts, abf1-1) alleles as well as strains bearing one wild-type or ts and one galactose-inducible allele (ABF1/GAL-ABF1; abf1-1/GAL-ABF1) at three different temperatures during growth and sporulation as indicated. Log2 ratios and the percentiles are indicated on the x-axis in (b), (c) and (d). For more information about Abf1 see reference (31).
Finally, we have integrated the regulatory motif prediction data published by Harbison et al. (33). The motifs are available via the ‘Features’ popup menu within the graphical display located in the ‘Transcripts’ section.
Protein interaction and network information
Whenever available, protein network information is organized into three classes that comprise nine categories: genetic (Dosage Lethality, Phenotypic Enhancement, Synthetic Lethality), in vivo (Affinity Capture-MS, Affinity Capture-Western, Co-purification, Two-hybrid) and in vitro (Biochemical Activity, Reconstituted Complex). Users can select each category individually or choose the option to display all interactions at once. This approach keeps the display simple and easy to read (Figure 3). For each interacting factor the gene name and annotation information as well as the references including a hyperlink to the respective PubMed entries are displayed. The gene names are hyperlinked with their corresponding report pages within GermOnline. Interaction data are provided by the BioGRID (18), and IntAct (19) resources.
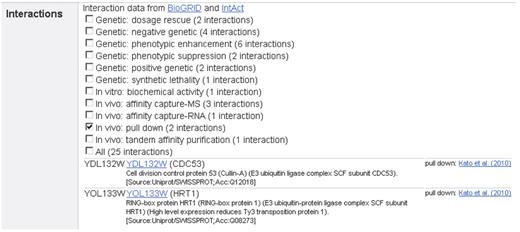
Protein interaction data display. Different interaction data available for Ndt80 are shown as an example. For clarity, users can choose to display only proteins found using a specific method, such as pull-down or select all interactions. Hyperlinks lead to the corresponding GermOnline gene report page and PubMed entries.
User access and community annotation
The first release of the database has been online from 2001 onwards as a web supplement for mitotic and meiotic yeast expression data (30,34). The second release including other species than yeast as well as a submission and curation system became available in 2003 (11–13). Since January 2007, when the third release was published (14), we have been using AWStats (http://sourceforge.net/projects/awstats/), a freely available tool that generates advanced web server statistics to monitor GermOnline access. The data are corrected for hits generated by computer programs or in the case of special hypertext transfer protocol status codes. During 2007–09 34 447 unique visitors have carried out 56 006 visits consulting 686 413 pages. In parallel, we monitored web traffic at GermOnlineWiki for two years and detected 9607 unique visitors carrying out 18 701 visits. During this period of time none of the wiki pages available were modified and no additional page was established. We conclude that while existing wiki entries are met with interest, the members of the community are not willing to edit and update them let alone establish new ones.
Utility and discussion
GermOnline 4.0 is a comprehensive online source of information about the expression of protein-coding genes relevant for the mitotic and meiotic cell cycle as well as germline development across selected species. The database portal now also provides DNA strand-specific tiling array expression data on yeast protein-coding and non-coding transcripts as well as information from yeast protein–DNA binding and protein–protein interaction studies. This sets the stage for future integration of similar data sets from experiments with mammalian samples.
The process of biocuration
Community annotation has been touted as an efficient approach to making scientific knowledgebase entries more complete and accurate (15). To provide an appropriate infrastructure for researchers working in the field of sexual reproduction we initially conceived a system based on controlled annotation vocabulary and a peer-review like approach (13). This was feasible but ultimately unsustainable because the annotation procedure was too complicated and most contributions involved more than one submission and curation cycle, which further increased the effort (and the cost) necessary to produce an entry of acceptable quality. We therefore switched to a simpler method based on the Wiki concept where external users establish and edit entries using free text and a single-step registration procedure without any review by an editorial board (14). However, the community failed to embrace the concept which in principle is also used by WikiGenes (35), WikiProteins (36) and GeneWiki, a portal within Wikipedia that aims at describing all human genes (37). These projects, like our own, all have one problem in common: writing a database entry does not in general count as a sign of productivity for a life scientist and does usually not help getting promoted or obtain funding. Therefore, any effort to establish meaningful and sustainable community annotation may need new conventions for the production and dissemination of scientific data (38–40). We note that a recent survey of 50 biologists who published data on genes or proteins suggested that active solicitation by biocurators and intuitive annotation tools together with concise guidelines would motivate them to participate in genome annotation (41). The survey also indicated that socio-economic factors which typically motivate life scientists to carry out research do not play a major role in this decision. While we have not conducted a formal survey of our users’ and contributors’ motivation (or lack thereof) our experience over several years does not seem to concur with this view. It is conceivable, however, that the perceived impact, importance and sustainability of a project may play an important role in the panoply of considerations that motivate potential community biocurators. In any case, our experience indicates that a project aiming at building a wiki for a specific biological process may experience difficulties no matter what type of interface researchers are being offered to work with.
Future work on the GermOnline portal
Keeping pace with swift technological development at the level of genome biological data production as well as within the field of bioinformatics is a vexing problem. In order to minimize the cost and workload required to provide users with the output of classical as well as emerging technologies we have decided to pursue a two-pronged strategy. We will maintain the current version of GermOnline based on Ensembl release 50 and MIMAS 3.0 such that classical 3′-UTR GeneChip data remain available even as this type of microarray technology (but not the data produced with them) becomes obsolete. In parallel, we are developing custom-tailored solutions such as the SGV for work based on novel and emerging research tools.
Our own laboratory and others are currently employing three major methods to study the transcriptome. Two of them are based on all-exon and tiling arrays which contain probes for all known exons (42) or the entire genome (9) of a given organism, respectively. The third one employs ultra-high throughput cDNA sequencing (UHTS) to determine the structure and the concentration of RNAs (RNA-Seq) (43). We intend to further develop genome-wide transcript viewers adapted to each technology. This likely includes moving from pre-computed images to a display generated on demand which requires substantial computing resources but has the key advantage of enabling us to hyperlink individual genes with external sources of information. Moreover, since recent advances in proteomics will likely lead to the production of quantitative expression data for most proteins (at last in the case of an organism such as budding yeast) in many conditions and strains we intend to include such data into the viewer to complement information on RNA (44).
Conclusion
GermOnline 4.0 is a cross-species gateway with a focus on genomics data for germline development, reproduction and cancer. Our database project has been continuously available to the community since 2001. From 2007 onwards, GermOnline has been based on Ensembl, MIMAS and MediaWiki. We have now focused the project’s mission on the dissemination of complex high-throughput expression data sets. As a consequence, a newly integrated prototype solution, SGV, is enabling life scientists, biomedical researchers and biocurators to work online with complex data sets providing information on expression levels and transcript structure across many experimental conditions.
Funding
Swiss Institute of Bioinformatics (to M. P.); bioinformatics platform of Biogenouest; Institut National de Santé et de Recherche Médicale, Young Investigator fellowship (to A. L.); University of Rennes 1 (No 913R310 to O. C.). Funding for open access charge: Inserm Avenir grant (No R07216NS to M.P.).
Conflict of interest. None declared.
Acknowledgements
We thank the members of the community for contributing their data. A.L. processed the tiling array expression data and developed the SGV, A.G. developed the database and server based on Ensembl and MIMAS, O.C. carried out database and infrastructure maintenance, F.C. contributed to tiling array expression data processing and SGV development, M.P. contributed to database design and wrote the paper. All authors read and approved the final article.
References
Author notes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.



