-
PDF
- Split View
-
Views
-
Cite
Cite
Andrei L. Turinsky, Sabry Razick, Brian Turner, Ian M. Donaldson, Shoshana J. Wodak, Literature curation of protein interactions: measuring agreement across major public databases, Database, Volume 2010, 2010, baq026, https://doi.org/10.1093/database/baq026
Close - Share Icon Share
Abstract
Literature curation of protein interaction data faces a number of challenges. Although curators increasingly adhere to standard data representations, the data that various databases actually record from the same published information may differ significantly. Some of the reasons underlying these differences are well known, but their global impact on the interactions collectively curated by major public databases has not been evaluated. Here we quantify the agreement between curated interactions from 15 471 publications shared across nine major public databases. Results show that on average, two databases fully agree on 42% of the interactions and 62% of the proteins curated from the same publication. Furthermore, a sizable fraction of the measured differences can be attributed to divergent assignments of organism or splice isoforms, different organism focus and alternative representations of multi-protein complexes. Our findings highlight the impact of divergent curation policies across databases, and should be relevant to both curators and data consumers interested in analyzing protein-interaction data generated by the scientific community.
Database URL:http://wodaklab.org/iRefWeb
Introduction
A myriad of cellular processes are carried out by groups of physically interacting proteins, or complexes, and the function of individual proteins often depends on their interaction partners. Substantial efforts are therefore being devoted worldwide to experimentally characterizing protein–protein interactions (PPIs) (1–9). This has in turn prompted the development of a number of specialized databases that curate and archive PPI data from the scientific literature and make them available to the scientific community (10).
Major PPI databases created in recent years such as HPRD (11), BioGRID (12) and IntAct (13), represent essentially independent annotation efforts driven by different research interests, and contain as a result complementary as well as redundant information. But exactly how much information is shared by the different databases and how much is unique, is generally not well documented, because comparing and integrating PPI information across the databases remains a challenging undertaking. The different databases apply different rules for capturing the data and often use different systems for cross-referencing genes and proteins across biological databases. Curation of the same publication by two different databases may hence result in significant discrepancies between the data that they record.
Adoption of the Proteomics Standards Initiative—Molecular Interaction (PSI-MI) controlled vocabulary and data structure (14) has been a major step forward in creating a common framework for representing PPI data. But although all major PPI databases adhere in principle to the PSI-MI standard, the actual implementations are still far from uniform. The outstanding differences prompted the creation of the IMEx consortium, committed to further unifying the PPI data representations and curation policies (15).
These standardization efforts have significantly eased the bottleneck for creating ‘meta’ resources that aggregate information from multiple PPI databases, with several such resources developed recently (16–20). But the aggregated PPI data made available by these resources are only partially normalized at best, due to many outstanding issues.
A number of problems continue to plague the curation and integration of PPI data. One problem, which further complicates the tedious task of assigning and cross-referencing gene and protein identifiers, is the annotation of protein isoforms. In some cases an interaction is specific to a particular protein isoform, whereas in others it is not. So far this information is rarely provided in the original publication. As a result the same protein may be annotated by two different databases as interacting with two different protein isoforms, each represented by a distinct identifier. Addressing this issue would require mapping the different isoforms of a protein to the corresponding gene (or ‘canonical’ isoform) (21), which is generally, but not always, uniquely defined. But this is currently not the accepted practice.
Guidelines on recording the organism in which an interaction has been observed also tend to differ. Some databases make the deliberate choice to curate only interactions pertaining to a specific organism from a given publication, or to infer interactions in a given organism (mainly human) on the basis of reported interactions in one or more related organisms (e.g. other mammals such as mouse or rat) (11,22). Problems with interpreting the published text are certainly also a factor, especially in studies of interactions in human, mouse or rat models. Indeed, cell lines from various model organisms are often used to draw conclusions about human cells. It is also not uncommon for authors to refer to previous studies for the description of cell lines and organisms, leaving it to the curator to trace earlier publications and resolve ambiguities.
The representation of multi-protein complexes identified by various detection methods (3,23,24) is yet another area where curations diverge. Complexes can be recorded either as a group of three or more associated proteins, or as a series of binary associations, depending on the practices adopted by the database (Supplementary Discussion S1). A common representation is the so-called spoke model, in which one protein is designated as a hub (or ‘bait’) and the complex is represented by a set of binary associations, each linking the bait to one of the other proteins (‘prey’) (25). Such associations may be distinguished from experimentally detected binary interactions (2,26) by examining the PSI-MI ‘interaction type’ record, since binary interactions derived from complexes are usually annotated as ‘association’ or ‘physical association’ (rather than ‘direct interaction’).
Databases also tend to differ on how they curate data sets produced by large-scale (high throughput) studies either on binary interactions or complexes. These studies often make available a high-confidence subset of the data, in addition to providing access to the full processed data set, or to the raw unprocessed data (2,27), but there is no general agreement between databases on which of these data sets is best fit for redistribution.
While all these problems are well known to database curators (28–30), the extent to which the ensuing differences impact the data currently stored across major PPI databases has so far not been quantified. The increasing number of non-experts who rely on PPI data for their research therefore often tend to ignore these problems altogether.
In this article we perform a quantitative evaluation of the level of agreement of the PPI data curated by major public databases. Our analysis is carried out on the global landscape of PPI data consolidated from nine major databases that focus primarily on the curation of experimentally derived physical PPIs: BIND (31), BioGRID (12), CORUM (22), DIP (32), IntAct (13), HPRD (11), MINT (33), MPact (34) and MPPI (35). The consolidation was performed using the Interaction Reference Index process (18) (‘Methods’ section), and data analysis was enabled by iRefWeb (http://wodaklab.org/irefweb), a web resource that serves as portal to the consolidated information (36).
The global PPI landscape with all its supporting evidence was generated by iRefIndex version 6.0. It comprised 271 716 distinct physical interactions involving 70 449 proteins. These interactions are associated with 1324 different organism-taxonomy identifiers and supported by a total of 42 651 publications. Interactions inferred by computational methods (37,38) and genetic interactions, which represent phenotype alterations produced by the mutation/deletion of one gene in the background of a mutation/deletion of another gene (39–41), were not considered here mainly because only a small subset of the databases curate them.
To perform the evaluation, we compared the annotations derived from the same publication by different databases. Whenever two databases cite the same publication as supporting an archived interaction, we used a similarity score to quantify the agreement between, respectively, the interactions and the proteins described in the original curated records, as outlined in Figure 1. These are two basic descriptors that (in principle) uniquely define the biological entity that was annotated. Ideally, they must be specified unambiguously by the curator and can be readily analyzed programmatically.
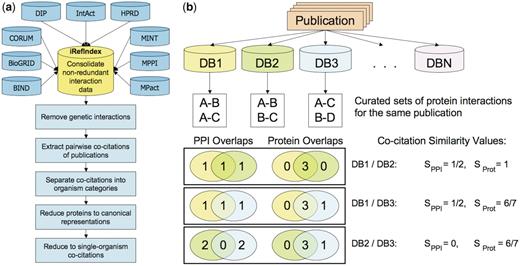
Pictorial overview of the analysis of pairwise co-citations of protein–protein interactions by different source databases from individual publications. (a) Workflow diagram summarizing the major steps of the co-citation analysis. A co-citation is defined as an instance of two databases citing the same publication in a protein interaction record. The first step is the consolidation of the PPI data from the nine databases analyzed in this work, performed by the iRefIndex procedures. Next, genetic interactions defined as described in ‘Methods’ section, are removed, and pairwise co-citations of individual publications by the source databases are extracted. Analysis is then performed on the bulk of these co-citations, as well as on co-citation subsets corresponding to publications dealing with interactions in one or more specific organisms (organism categories), in only a single specific organism (single-organism), and after systematically mapping proteins to their canonical isoforms (canonical representation) (see text). (b) Evaluating the consistency in pairwise co-citations of a hypothetical publication cited by three databases out of the total of nine analyzed here. Sorensen–Dice similarity scores (Methods section) are computed for each pairwise co-citation, to quantify overlaps between the sets of interactions (SPPI) and proteins forming these interactions (SProt). The distributions of these quantities are then used to evaluate the level of consistency in different co-citation categories.
Analysis of the 15 471 shared publications reveals that on average, two databases fully agree on only 42% of the interactions and 62% of the proteins curated from the same publication, but the level of agreement for individual publications varies considerably. We then quantify how this initial level of agreement is globally impacted by factors such as divergent annotation of organisms, different splice isoform assignments and alternative representations of multi-protein complexes. Our findings highlight the role played by differences in curation policies (past or present) across databases. They also underscore the challenges that annotators face in interpreting published information and provide valuable insight into the hurdles that bioinformaticians need to overcome to integrate PPI data from multiple sources. Our study should help in formulating recommendations and developing improved software tools for all those interested in recording, integrating and analyzing protein interaction data.
Results
Level of agreement across databases on a per-publication basis
In order to measure the agreement of the information curated across databases on a per-publication basis, we examine the subset of shared publications, i.e. those curated by two or more databases. We define a ‘co-citation’ as an instance of two databases citing the same publication in an interaction record (‘Methods’ section). Depending on the number of curating databases, a single publication may give rise to several pairwise co-citations (Figure 1). Only ∼36% of all cited publications, numbering 15 471, are shared, and those give rise to 27 399 pairwise co-citations.
For each pairwise co-citations we compute two similarity scores, SPPI and SProt, which take values between 0 and 1 and measure the agreement, respectively, between the annotated PPIs, and between the annotated proteins engaged in these interactions (‘Methods’ section). Figure 2a plots the distributions of the similarity scores for the interactions (horizontal axis) and for the set of annotated proteins (vertical axis). It shows that the level of agreement between the annotated information in the analyze co-citations, ranges between full agreement and complete disagreement. On average, two databases curating the same publication agree on 42% of their interactions. The discrepancies between the sets of proteins annotated from the same publication are typically less pronounced, with the average agreement of 62%, but the overall trend is similar.
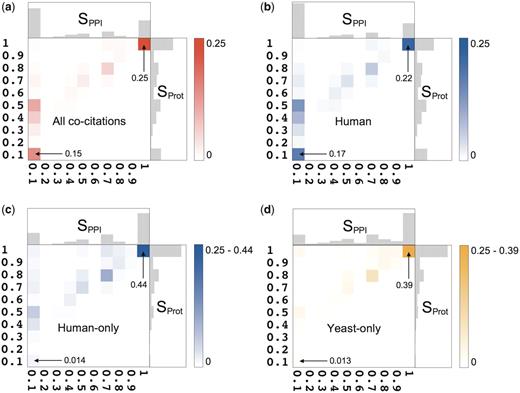
Statistical summary of the pairwise co-citation landscape across nine source databases. (a) Two-dimensional frequency distribution of Sorensen–Dice scores (given as fractions) for interactions (horizontal axis, SPPI) and proteins (vertical axis, SProt), over all co-citations. The color scale indicates the frequency. One-dimensional distributions of these scores are shown along the corresponding axes. The mean and standard deviation of SPPI are 0.42 ± 0.42, hence two databases curating the same publication agree (on average) on only 42% of the interactions. Both databases record identical sets of interactions (SPPI = 1) in 24% of co-citations, while in 42% of co-citations they record completely different PPIs (SPPI = 0). The remaining 34% represent partial agreement, varying widely between the two extremes. The mean and standard deviation of SProt are 0.62 ± 0.35. Full agreement (SProt = 1) occurs in 29% of co-citations, a comparable level to that obtained for interactions, whereas complete disagreement (SProt = 0) occurs in only 14% of the cases, or almost three times less frequently than for interactions. (b) The two-dimensional frequency distribution of Sorensen–Dice scores in the Human category, i.e. 20 671 co-citations in which at least one database recorded human proteins. (c ) Two-dimensional frequency distribution of the Sorensen–Dice similarity scores for the 15 194 human-only co-citations. Despite a prominent peak near the perfect agreement (SPPI = 1/SProt = 1), ∼57% of the co-citations display various levels of partial agreement. (d) Distribution of the Sorensen–Dice similarity scores for the 4983 yeast-only co-citations. Despite a prominent peak at the perfect agreement, ∼64% of the co-citations display various levels of partial agreement.
Admittedly, our criterion for agreement is quite strict, as it requires that the two databases refer to the exact same amino acid sequence and same organism taxonomy identifier when annotating each interacting protein. However, this allows us to quantify how far we are from the ideal case of perfect agreement at the starting point of our investigation. Seeing that the levels of agreement on both the annotated interactions and proteins are low, we now examine some of the factors that contribute to the observed differences.
Publications dealing with specific organisms
Since the experimental characterization of PPIs tends to vary between organisms in terms of both the methodology and coverage, we investigate how the level of agreement varies across publications dealing with specific organisms. Organism information is usually unambiguously recorded using the NCBI taxonomy identifiers (42) and can therefore be readily analyzed and compared. We consider a co-citation as pertaining to a given organism when at least one of the two databases citing the corresponding publication recorded proteins from that organism. All 27 399 co-citations are classified in this fashion into 1324 categories corresponding to specific organisms. In this classification a given co-citation can belong to more than one organism category, because the same publication may be interpreted differently by two databases, with for example, one database recording a human interaction but the other recording an interaction from mouse.
The organism categories vary widely in the number of co-citations that they contain and in the agreement levels of the co-citations therein. The comparison across categories is summarized in Figure 3a, which plots the SPPI and SProt scores for categories with at least 50 co-citations. Among the largest categories, co-citations corresponding to yeast Saccharomyces cerevisiae stand out as displaying the highest agreement level (SPPI and SProt averaging 63 and 80%, respectively), whereas co-citations dealing with mouse or rat display very poor agreement (with average SPPI and SProt being, respectively, 12 and 26% for mouse and 11 and 26% for rat). The average agreement for human co-citations is roughly in the middle of the range (37, 58%). For other well-studied organisms such as fission yeast Schizosaccharomyces pombe, plant Arabidopsis thaliana, worm Caenorhabditis elegans and fly Drosophila melanogaster, the average agreement is relatively high, whereas in some of the vertebrate species and in the bacteria Escherichia coli it is significantly lower (Figure 3a).
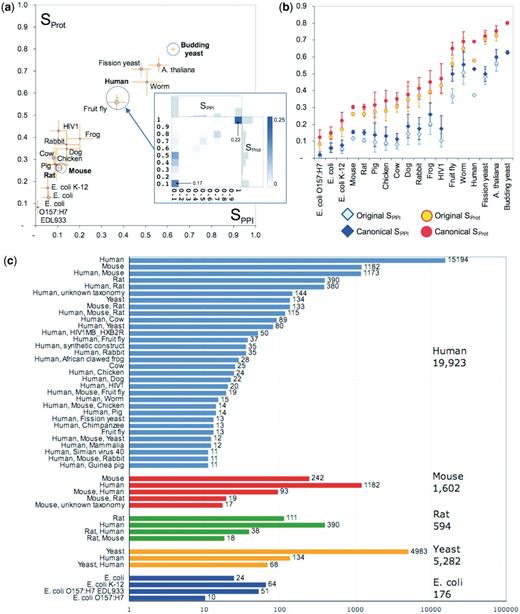
Analysis of co-citation agreement within different organism categories. (a) Average Sorensen–Dice similarity score for co-citations in the different organism categories, before the canonicalization of protein isoforms. The area of the circle surrounding the data point is proportional to the number of co-citations within the category. Orange error bars indicate the 95% confidence interval for each category’s mean. Only organisms with at least 50 co-citations are shown. The four largest categories are Human (20 671 co-citations), the yeast S. cerevisiae (5444 co-citations), Mouse (3550 co-citations) and Rat (R. norvegicus, 1477 co-citations). The inset shows the two-dimensional similarity distribution for the Human category (same as in Figure 2b). (b) Improvement in average similarity scores for organism categories upon mapping of proteins to their canonical splice isoforms. Error bars indicate the 95% confidence interval for each category’s mean. Improved agreement is observed for human and fly co-citations, and to a lesser extent for the mouse and rat co-citations. The small improvements observed for E. coli co-citations are due to a more consistent strain assignment performed in parallel to the canonical isoform mapping. (c) Discrepancies in organism assignments: each group of colored bars corresponds to co-citations in which one database records proteins from a single organism (indicated on the right, with the total number of such citations). Each colored bar represents co-citations in which the other database records the organisms indicated on the right. Only bars with at least 10 co-citations are shown.
Divergent isoform assignments
The relatively low agreement level within the vertebrate categories led us to examine the extent to which the detected discrepancies were affected by differences in splice-isoform assignments. We therefore compared the level of agreement across the PPI landscape before and after the splice isoform normalization process (‘Methods’ section). This process reduced the original set of 271 716 interactions involving 70 449 proteins, to that of 248 465 interactions involving 63 871 proteins. At the same time it increased the level of agreement from 42 to 54 ± 41% for PPIs, and from 62 to 71 ± 33% for proteins. Considering specific organisms, the agreement improved for many species, especially for human and fly (Figure 3b). We thus confirm that, using different splice isoforms in the description of gene products is indeed a significant contributor to annotation discrepancies.
Following these findings, information provided by the iRefWeb interface refers to proteins and interactions mapped to their canonical isoforms (36). However, information on the particular splice isoforms curated by the sources databases is preserved and can be queried, as each consolidated interaction links back to the original records from which is was derived.
Divergent organism assignments
Even after the consolidation of isoforms, the level of agreement associated with major organism categories, such as Mouse and Rat, remained <20% for the interactions and <30% for proteins (Figure 3b). To probe further into the impact of organism assignments, we investigated the following question: When two databases cite the same article, and one of them records all proteins as belonging to organism A, then which organism(s) does the other database record, and how often?
Results show that the disagreements on the annotated organism and annotation of PPIs from different organisms by different databases are quite common (Figure 3c). The discrepancies for the Mouse or Rat categories are dramatic. Of the 1602 co-citations in which one database records exclusively mouse proteins, the other database does so in only 242 cases (15%). Most commonly, however (in 1182 cases, or ∼74%), human proteins are recorded instead. The trend is very similar for the smaller Rat category. For the category Human, there are 19 923 pairwise co-citations in which one of the two databases records exclusively human proteins. In 4729 of these cases (or 24%) the other database reports proteins from a different organism (most often mouse or rat) or a combination of organisms.
In contrast, organism assignments are much more consistent for S. cerevisiae, A. thaliana, S. pombe, worm and fly (Figure 3c), with only rare instance where interactions of other organisms (mainly human) are recorded instead. Additionally, discrepancies for the fission yeast, S. pombe, often involve the attribution of proteins to the yeast S. cerevisiae by the other database. Discrepancies are also observed for the E. coli category, but are mostly a result of different annotations of E. coli strains (Figure 3c).
In order to factor out the effects of divergent organism assignments, we further restrict our analysis to co-citations in which both databases record proteins from the same organism. In such co-citations improvements in annotation consistency of the order of 20% are observed for several organism categories (Table 1 and Figure 2c and d), confirming that divergent organism assignments contribute significantly to the observed differences.
Agreement for the largest single-organism categories of pairwise co-citations
| Organism . | SPPI . | SProt . | SPPI P-value . | SProt P-value . | Pubs . | Co-cite . |
|---|---|---|---|---|---|---|
| Human | 0.66 (0.37) | 0.83 (0.22) | 0 | 0 | 10 546 | 15 194 |
| Yeast | 0.66 (0.35) | 0.84 (0.22) | 2.9e-4 | 3.6e-4 | 1867 | 4983 |
| Mouse | 0.42 (0.45) | 0.65 (0.34) | 0 | 0 | 203 | 242 |
| Arabidopsis thaliana | 0.63 (0.36) | 0.79 (0.24) | 0.486 | 0.306 | 156 | 186 |
| Fission yeast | 0.63 (0.33) | 0.85 (0.19) | 1.5e-3 | 2.9e-6 | 123 | 162 |
| Fruit fly | 0.66 (0.36) | 0.81 (0.23) | 5.6e-5 | 1.2e-6 | 106 | 147 |
| Rat | 0.53 (0.42) | 0.76 (0.27) | 0 | 0 | 95 | 111 |
| Worm | 0.70 (0.33) | 0.84 (0.18) | 0.0427 | 0.0189 | 19 | 37 |
| Escherichia coli | 0.32 (0.48) | 0.50 (0.40) | 0.044 | 9.9e-6 | 15 | 24 |
| Organism . | SPPI . | SProt . | SPPI P-value . | SProt P-value . | Pubs . | Co-cite . |
|---|---|---|---|---|---|---|
| Human | 0.66 (0.37) | 0.83 (0.22) | 0 | 0 | 10 546 | 15 194 |
| Yeast | 0.66 (0.35) | 0.84 (0.22) | 2.9e-4 | 3.6e-4 | 1867 | 4983 |
| Mouse | 0.42 (0.45) | 0.65 (0.34) | 0 | 0 | 203 | 242 |
| Arabidopsis thaliana | 0.63 (0.36) | 0.79 (0.24) | 0.486 | 0.306 | 156 | 186 |
| Fission yeast | 0.63 (0.33) | 0.85 (0.19) | 1.5e-3 | 2.9e-6 | 123 | 162 |
| Fruit fly | 0.66 (0.36) | 0.81 (0.23) | 5.6e-5 | 1.2e-6 | 106 | 147 |
| Rat | 0.53 (0.42) | 0.76 (0.27) | 0 | 0 | 95 | 111 |
| Worm | 0.70 (0.33) | 0.84 (0.18) | 0.0427 | 0.0189 | 19 | 37 |
| Escherichia coli | 0.32 (0.48) | 0.50 (0.40) | 0.044 | 9.9e-6 | 15 | 24 |
Mean and standard deviation (in parentheses) of the Sorensen–Dice SPPI and SProt distributions, considering only co-citations where both databases record proteins from the same organism, using canonical splice isoforms. Only a few single-organism categories remained large enough for meaningful analysis. P-values of the Kolmogorov–Smirnov test (‘Methods’ section) are shown in comparison to the overlapping organism categories in Figure 3b (in both cases after the canonical-isoform mapping of proteins was performed). The number of pairwise co-citations (‘Co-cite’) and publications that give rise to these co-citations (‘Pubs’) is also shown. The agreement for the Human-only and Fly-only categories now becomes as high as that for yeast S. cerevisiae. Several-fold improvements are observed for Mouse and Rat. The already-high agreement for Yeast shows little improvement. After the Bonferroni correction with α = 0.025, improvements for A. thaliana, Worm and E. coli are not statistically significant.
Agreement for the largest single-organism categories of pairwise co-citations
| Organism . | SPPI . | SProt . | SPPI P-value . | SProt P-value . | Pubs . | Co-cite . |
|---|---|---|---|---|---|---|
| Human | 0.66 (0.37) | 0.83 (0.22) | 0 | 0 | 10 546 | 15 194 |
| Yeast | 0.66 (0.35) | 0.84 (0.22) | 2.9e-4 | 3.6e-4 | 1867 | 4983 |
| Mouse | 0.42 (0.45) | 0.65 (0.34) | 0 | 0 | 203 | 242 |
| Arabidopsis thaliana | 0.63 (0.36) | 0.79 (0.24) | 0.486 | 0.306 | 156 | 186 |
| Fission yeast | 0.63 (0.33) | 0.85 (0.19) | 1.5e-3 | 2.9e-6 | 123 | 162 |
| Fruit fly | 0.66 (0.36) | 0.81 (0.23) | 5.6e-5 | 1.2e-6 | 106 | 147 |
| Rat | 0.53 (0.42) | 0.76 (0.27) | 0 | 0 | 95 | 111 |
| Worm | 0.70 (0.33) | 0.84 (0.18) | 0.0427 | 0.0189 | 19 | 37 |
| Escherichia coli | 0.32 (0.48) | 0.50 (0.40) | 0.044 | 9.9e-6 | 15 | 24 |
| Organism . | SPPI . | SProt . | SPPI P-value . | SProt P-value . | Pubs . | Co-cite . |
|---|---|---|---|---|---|---|
| Human | 0.66 (0.37) | 0.83 (0.22) | 0 | 0 | 10 546 | 15 194 |
| Yeast | 0.66 (0.35) | 0.84 (0.22) | 2.9e-4 | 3.6e-4 | 1867 | 4983 |
| Mouse | 0.42 (0.45) | 0.65 (0.34) | 0 | 0 | 203 | 242 |
| Arabidopsis thaliana | 0.63 (0.36) | 0.79 (0.24) | 0.486 | 0.306 | 156 | 186 |
| Fission yeast | 0.63 (0.33) | 0.85 (0.19) | 1.5e-3 | 2.9e-6 | 123 | 162 |
| Fruit fly | 0.66 (0.36) | 0.81 (0.23) | 5.6e-5 | 1.2e-6 | 106 | 147 |
| Rat | 0.53 (0.42) | 0.76 (0.27) | 0 | 0 | 95 | 111 |
| Worm | 0.70 (0.33) | 0.84 (0.18) | 0.0427 | 0.0189 | 19 | 37 |
| Escherichia coli | 0.32 (0.48) | 0.50 (0.40) | 0.044 | 9.9e-6 | 15 | 24 |
Mean and standard deviation (in parentheses) of the Sorensen–Dice SPPI and SProt distributions, considering only co-citations where both databases record proteins from the same organism, using canonical splice isoforms. Only a few single-organism categories remained large enough for meaningful analysis. P-values of the Kolmogorov–Smirnov test (‘Methods’ section) are shown in comparison to the overlapping organism categories in Figure 3b (in both cases after the canonical-isoform mapping of proteins was performed). The number of pairwise co-citations (‘Co-cite’) and publications that give rise to these co-citations (‘Pubs’) is also shown. The agreement for the Human-only and Fly-only categories now becomes as high as that for yeast S. cerevisiae. Several-fold improvements are observed for Mouse and Rat. The already-high agreement for Yeast shows little improvement. After the Bonferroni correction with α = 0.025, improvements for A. thaliana, Worm and E. coli are not statistically significant.
Specific examples of divergent organism assignments
Experiments on mammalian cells and proteins are of crucial importance to the studies of human diseases, especially when such experiments involve known disease-related proteins. However, disagreements between databases are common even in publications involving such proteins.
Figure 4a illustrates the difficulty of curating a publication describing interactions between a well-known breast cancer protein BRCA1 and another protein, BAP1, which binds to BRCA1 and enhances cell-growth suppression (43). The published text describes the interactions between the BRCA1 RING finger domain (which has the same sequence in human and mouse) with the human BAP1 protein as well as with different variants of the BAP1 mouse ortholog. The three databases that cite this study differ in their representation of BAP1 as either a human or a mouse interactor of BRCA1, with only IntAct faithfully representing both versions.
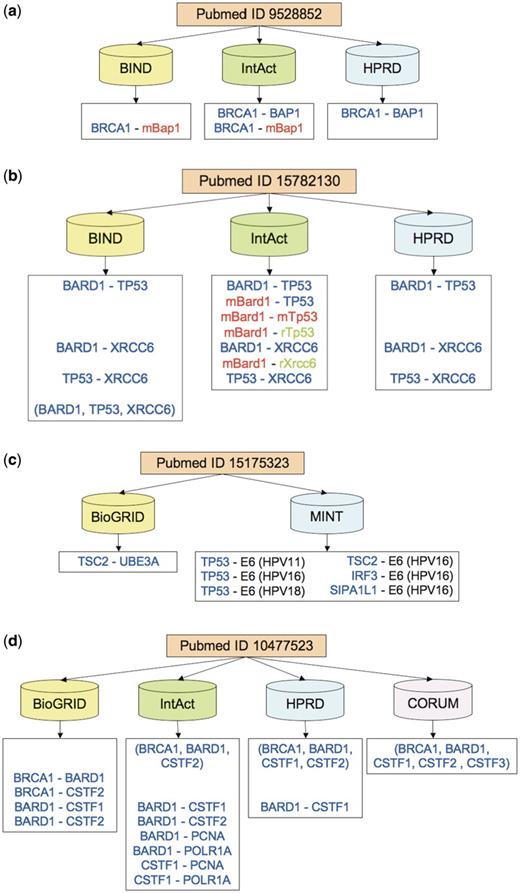
Examples of citation discrepancies. Protein colors indicate the organism (human in blue, mouse in red, rat in green). Prefixes ‘m’ and ‘r’ indicate mouse and rat, respectively. Matching pairwise interactions are aligned horizontally across databases. (a) In a study involving BRCA1 (breast cancer 1) protein, its interactor BAP1 (BRCA1 associated protein-1) is attributed to either human (by HPRD), or mouse (by BIND), or both (by IntAct). BIND and HPRD are in complete disagreement on interactions. (b) Three databases annotate a study involving TP53 (tumor protein p53), BARD1 (BRCA1 associated RING domain 1) and XRCC6 (X-ray repair complementing defective repair in Chinese hamster cells 6). BIND and HPRD record only human interactions, whereas IntAct also records PPI versions involving mouse and rat orthologs. BIND additionally annotates a human complex involving all three proteins. (c) Citing an article on insulin-pathway interference, MINT records interactions between human-papillomavirus (HPV) oncoprotein E6, which in implicated in cervical cancer, and several human proteins, including tumor suppressors TP53 and TSC2. In contrast, BioGRID cites the same study to support only one interaction, between TSC2 and a human ubiquitin protein ligase UBE3A related to the neuro-genetic Angelman syndrome. (d) Four databases record interactions between BRCA1, BARD1 and several cleavage stimulation factors (CSTF, subunits 1–3). All databases except BioGRID record a protein complex but disagree on its precise membership. All except CORUM also record various pairwise interactions of the type ‘physical association’ among BRCA1, BARD1 and CSTF1-2. In addition, IntAct records interactions with two additional proteins, PCNA and POLR1A. CORUM is in complete disagreement on interactions with the other three databases but in high agreement on the proteins involved.
Another publication describes the interactions of only three proteins, including a well-known tumor suppressor TP53 as well as a BRCA1-associated protein BARD1 (44). The experiments were conducted using human prostate-cancer cells, rat ovarian-cancer cells and proteins from human, rat and mouse. As a result, the difficulty of correctly interpreting the paper rises dramatically and the annotations have more potential to differ. Two of the databases record only human interactions, with BIND recording them both as a single complex comprising three proteins and three distinct pairwise association (Figure 4b). In contrast, curating the same paper, IntAct records interactions involving human, mouse and rat proteins.
The presence of multiple organisms in a PPI annotation is not in itself a sign of annotation discrepancies or difficulties, since some publications may report interactions between host and pathogen proteins. Figure 4c illustrates the annotations derived from such a study, which investigated the interference of the human papillomavirus (HPV) with the human insulin-signaling pathway (45). However, only one of the two databases annotates the interactions between human and HPV proteins, whereas the other database does not record even a single HPV protein interactor, for reasons that are not clear.
Several additional examples of annotation differences can be found in the Supplementary Discussion S3.
Other factors affecting protein identification
Even when both databases completely agree on the organism assignment, and after splice isoform normalization, agreement levels for interactions for on the largest organism categories except worm do not exceed 66% (Table 1). To elucidate the factors that contribute to the outstanding differences, we analyze the two largest categories of the human-only and yeast (S. cerevisiae)-only co-citations, which together represent the bulk (74%) of all 27 399 co-citations in our data.
First, we examine co-citations in which the two databases disagree on every PPI described in the publication (SPPI = 0). Such co-citations comprise 17% of the human-only and 13% of the yeast-only categories (Figure 2c and d). In most of these cases the two databases have a partial overlap between the annotated sets of proteins. This suggests that a major source of the remaining disagreements on PPIs, after splice isoform normalization has been performed, is the divergent identification of individual proteins by the databases. A protein may be specified differently due either to the existence of multiple representations that cannot be easily mapped to the same gene by our consolidation procedure, or to a genuine curation discrepancy. In such cases, the divergence propagates to the corresponding PPI records in the two databases, causing them to differ even if they agree on a fraction of the interacting proteins. We anticipate that this subset of the data is enriched for papers that should be re-examined by curators of the source databases for potential issues with curation errors or other genuine differences.
Treatment of binary interactions versus complexes
In a small fraction of the co-citations disagreements on the interactions persist despite complete agreement on the proteins involved. Indeed, 1% of all human-only and 2% of yeast-only co-citations agree perfectly on the annotated proteins (SProt = 1), but disagree completely on the reported interactions (SPPI = 0). The main origin of these disagreements is the group versus binary representations of multi-protein complexes, as already mentioned.
For example, Figure 4d details the curated information from an experimental study (46) that identified a protein complex of the breast cancer protein BRCA1, the BRCA1-associated protein BARD1, and a cleavage-stimulation factor CSTF. The four databases that annotated the paper record the complex differently, using either multi-subunit groups, or binary expansions, or both. However, they largely agree (with BioGRID and HPRD agreeing completely) on the sets of proteins involved in these interactions.
Overall, co-citations involving groups of proteins display significantly lower agreement on PPIs (29% for human-only and 34% for yeast-only co-citations, on average) than those that deal with binary representations (72 and 70% on average, respectively). However, the agreement on the proteins involved in multi-protein groups remains rather high (76 and 86% on average, respectively; Table 2).
| Organism . | DBs annotating complexes . | SPPI . | SProt . | Pubs . | Co-cite . |
|---|---|---|---|---|---|
| Human | None | 0.72 (0.35) | 0.84 (0.22) | 9737 | 13 327 |
| One | 0.27 (0.29) | 0.73 (0.22) | 867 | 1412 | |
| Both | 0.37 (0.37) | 0.84 (0.18) | 376 | 455 | |
| Yeast | None | 0.70 (0.34) | 0.84 (0.22) | 1690 | 4511 |
| One | 0.34 (0.31) | 0.85 (0.19) | 261 | 439 | |
| Both | 0.37 (0.37) | 0.91 (0.16) | 26 | 33 |
| Organism . | DBs annotating complexes . | SPPI . | SProt . | Pubs . | Co-cite . |
|---|---|---|---|---|---|
| Human | None | 0.72 (0.35) | 0.84 (0.22) | 9737 | 13 327 |
| One | 0.27 (0.29) | 0.73 (0.22) | 867 | 1412 | |
| Both | 0.37 (0.37) | 0.84 (0.18) | 376 | 455 | |
| Yeast | None | 0.70 (0.34) | 0.84 (0.22) | 1690 | 4511 |
| One | 0.34 (0.31) | 0.85 (0.19) | 261 | 439 | |
| Both | 0.37 (0.37) | 0.91 (0.16) | 26 | 33 |
Values for the Sorensen–Dice distributions [mean and standard deviation (in parentheses)] for shared publications (co-citations) are computed after the mapping of proteins to their canonical splice isoforms. ‘None/One/Both’ indicate pairwise co-citations where, respectively, neither of the DBs represents multi-protein complexes as groups of proteins, only one DB uses the group representation, and both DBs use that representation. The number of pairwise co-citations of publications in each category is shown (‘Co-cite’), along with the number of shared publications (‘Pubs’) that give rise to these co-citations. As expected, co-citations where at least one database uses the group representation, display significantly lower SPPI values on average, than those that do not use it. However, they feature high-average SProt values, indicating a significantly better agreement on the proteins involved than on their grouping into interactions.
| Organism . | DBs annotating complexes . | SPPI . | SProt . | Pubs . | Co-cite . |
|---|---|---|---|---|---|
| Human | None | 0.72 (0.35) | 0.84 (0.22) | 9737 | 13 327 |
| One | 0.27 (0.29) | 0.73 (0.22) | 867 | 1412 | |
| Both | 0.37 (0.37) | 0.84 (0.18) | 376 | 455 | |
| Yeast | None | 0.70 (0.34) | 0.84 (0.22) | 1690 | 4511 |
| One | 0.34 (0.31) | 0.85 (0.19) | 261 | 439 | |
| Both | 0.37 (0.37) | 0.91 (0.16) | 26 | 33 |
| Organism . | DBs annotating complexes . | SPPI . | SProt . | Pubs . | Co-cite . |
|---|---|---|---|---|---|
| Human | None | 0.72 (0.35) | 0.84 (0.22) | 9737 | 13 327 |
| One | 0.27 (0.29) | 0.73 (0.22) | 867 | 1412 | |
| Both | 0.37 (0.37) | 0.84 (0.18) | 376 | 455 | |
| Yeast | None | 0.70 (0.34) | 0.84 (0.22) | 1690 | 4511 |
| One | 0.34 (0.31) | 0.85 (0.19) | 261 | 439 | |
| Both | 0.37 (0.37) | 0.91 (0.16) | 26 | 33 |
Values for the Sorensen–Dice distributions [mean and standard deviation (in parentheses)] for shared publications (co-citations) are computed after the mapping of proteins to their canonical splice isoforms. ‘None/One/Both’ indicate pairwise co-citations where, respectively, neither of the DBs represents multi-protein complexes as groups of proteins, only one DB uses the group representation, and both DBs use that representation. The number of pairwise co-citations of publications in each category is shown (‘Co-cite’), along with the number of shared publications (‘Pubs’) that give rise to these co-citations. As expected, co-citations where at least one database uses the group representation, display significantly lower SPPI values on average, than those that do not use it. However, they feature high-average SProt values, indicating a significantly better agreement on the proteins involved than on their grouping into interactions.
Representing complexes as groups of proteins, is currently not a widely adopted practice, and occurs in just 12% of the human-only and 9% of yeast-only co-citations. However, it accounts for, respectively, 32 and 22% of co-citations showing complete disagreement on PPIs (SPPI = 0). Furthermore, it accounts for as many as 75% of the human-only and 45% of yeast-only co-citations showing complete disagreement on PPIs but full agreement on the proteins involved (SPPI = 0, SProt = 1). These numbers do not include co-citations where both databases translated the protein groups (complexes) into sets of binary interactions, which we could not systematically trace, and which we suspect may contribute to some of the remaining cases of low SPPI, mainly due to different conventions used to perform the translation.
Contribution of specific databases to the observed trends
Different pairs of databases display different levels of agreement in each organism category. The average pairwise agreement on PPIs between the source databases, and the corresponding number of co-citations for the human-only and yeast-only co-citations, respectively, are summarized in Figure 5, and the Supplementary Tables S3 and S4.
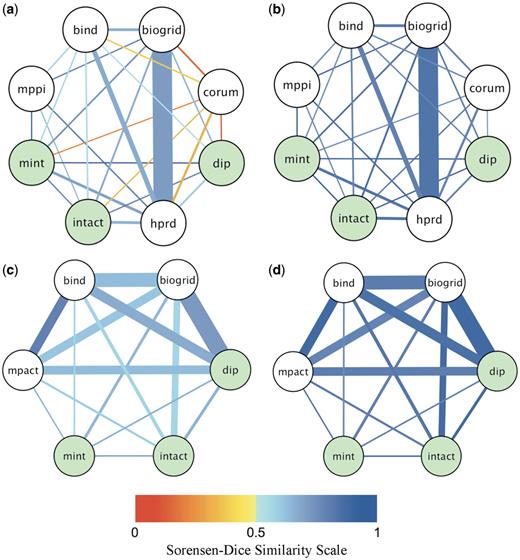
Pairwise agreement between databases for yeast-only and human-only co-citations. Shown is a pictorial summary of the agreement levels between pairs of databases for shared publications, where both databases annotated all the interactions reported in the shared publication to the same organism. The thickness of the edge connecting two databases is proportional to the fraction of the total number of shared (co-cited) publications contributed by the database pair. The edge color indicates the value of the average Sorensen–Dice similarity coefficient according to the color scale shown at the bottom (shades of orange for agreement on less than half of the interactions or proteins, shades of blue for agreement on more than half of interactions or proteins). (a) Fraction of co-citations and agreement on interactions (SPPI) for human-only co-citations. (b) Fraction of co-citations and agreement on proteins (SProt) for human-only co-citations. (c) Fraction of co-citations and agreement on interactions (SPPI) for yeast-only co-citations. (d) Fractions of co-citation and agreement on proteins (SProt) for yeast-only co-citations. The Human-only data set is dominated by co-citations from BioGRID and HPRD, whereas the overlap in yeast-only citations is contributed more evenly by most databases except MINT. The levels of agreement are markedly improved, compared to those observed in all co-citations, before and after the canonicalization of splice isoforms (Supplementary Figure S1). The agreement on proteins is overall better that the agreement on interactions for each database pair. Persistent differences are found in co-annotations involving CORUM (22), which annotates mammalian complexes: the average Sorensen–Dice similarity score for CORUM and any other source database is below 0.5, primarily due to different representations of complexes (Supplementary Discussion S1). Green nodes correspond to IMEx databases (DIP, IntAct, MINT). Although their agreement levels are somewhat higher than average for human-only co-citations, they represent only 1% of all human-only and 3.7% of all yeast-only co-citations analyzed here. Additional details are provided in the Supplementary Tables S3 and S4.
These summaries show that as many as 8340 out of 15 194 human-only co-citations (or 55%) are those by BioGRID and HPRD (Figure 5a and b and Supplementary Table S3). The next largest overlap is the 1653 co-citations (11%) by BIND and HPRD, with all other overlaps not exceeding 5%. Naturally, the average agreement level between BioGRID and HPRD (SPPI = 0.71, SProt = 0.84) prominently affects the distribution of the similarity values for human data. Most other pairs of databases have somewhat lower, but comparable agreement levels, with the exception of CORUM, which annotates mammalian protein complexes. CORUM participates in 1018 human-only co-citations, displaying a low average agreement on PPIs (SPPI = 0.27 ± 0.38) but a high agreement on proteins (SProt = 0.78 ± 0.23) with the other databases. This low level of agreement stems from the fact that the CORUM database represents complexes as groups of associated proteins. Such group representations will invariably display disagreements with binary expansions derived from the same published information, due to differences in protein composition. Also, two group representations independently curated from the same article are, in general, less likely to have identical protein compositions, further contributing to the observed differences.
The overlap between databases for yeast-only shared publications is distributed more evenly than for the human-only articles (Figure 5c and d and Supplementary Table S4). Of the 4983 yeast-only co-citations, 21% are those by BioGRID and DIP, 17% by BioGRID and BIND, 12% by BIND and DIP, 11% by BIND and MPact, 10% by DIP and MPact, etc. As in the case of human data, the average agreement rates across different database pairs are similar, especially for pairs with a significant overlap in cited publications.
Clearly the pairwise agreement levels for these organism-specific co-citations are significantly higher than those obtained for co-citations prior to factoring out divergent protein representation and/or organism assignments. Indeed, for the latter type of co-citations many pairs of databases agree on less than half of PPIs on average (Supplementary Table S1 and Supplementary Figure S1).
Interestingly, the pairwise agreement between members of the IMEx databases (DIP, IntAct, MINT) is in general better than average, albeit similar to those of some other database pairs. This is observed both before and after elimination of some of the major discrepancy-causing factors (Figure 5, Supplementary Figure S1 and Supplementary Data), but is unlikely to fully reflect the common curation policies adopted by this consortium, since much of the data currently stored in the IMEx databases predates the implementation of these policies.
Discussion
In this study we quantified the level of agreement in the PPI data curated from 15 471 publications co-cited across nine major public databases. In doing so we evaluated the global impact of several factors on the consistency of the curated information.
One key factor is the divergence in organism assignments, which was detected in 21% of all co-citations (5769 out of a total of 27 399 co-citations). This divergence may sometimes results from the difficulty of interpreting the complex information reported in the original publication. Most commonly, however, it is due to the application of different curation rules, with some databases recording only interactions in the organism of interest, or systematically transferring interactions identified in one organism to its orthologs in another. Differences in organism assignments across databases should be dramatically reduced by adherence to common curation policies that would, for example, stipulate flagging interactions inferred by homology or impose stricter rules for selecting publications to curate.
Another factor, which contributes significantly to the detected differences, is the treatment of multi-protein complexes. Of the 3470 co-citations that involve complexes, only 76 are in complete agreement following the normalization of splice isoforms, indicating that up to 3394 (or 12% of the full data set) might be affected by this factor. As already mentioned, this poor agreement level is mostly due to different representations of the data that cannot be readily inter-converted (Supplementary Discussions S1 and S3). Adopting a common convention according to which multi-protein complexes identified by various purification methods are represented as groups of associated proteins (15) is a simple solution that should significantly improve agreement levels. However, once such convention is widely adopted, the criteria for quantifying the agreement between two databases curating the same reported complex should be relaxed from requiring a perfect match between the annotated proteins, as done here, to quantifying the level of overlap between the two protein lists.
By far the most crucial factor affecting the agreement levels analyzed here is the proper assignment of protein and gene identifiers across biological databases. Our analysis relies completely on the iRefIndex consolidation procedure for establishing the identity of the proteins in the PPI records. This procedure maps the variety of protein identifiers recorded by the databases to the protein amino acid sequences, using a series of steps (18). The last step, introduced recently, maps proteins to their canonical splice-isoforms (or corresponding genes).
Here we were able to evaluate the contribution to the observed disagreements both before and after this last mapping step. Our isoform normalization method was able to eliminate all disagreements on proteins in 2675 co-citations, or nearly 10% of the co-citation data set. It increased the fraction of co-citations with a perfect agreement on proteins from 29 to 39%, and those with perfect agreement on interactions from 24 to 32%.
The problem of cross-referencing proteins and genes across biological databases is an endemic one, over which the PPI databases have very limited control. Addressing it in the context of PPI curation needs to involve the cooperation of database curators, bioinformaticians as well as the authors of experimental studied (47,48). With this goal in mind, concrete proposals on how to help authors of publications provide standardized descriptions of interactions have recently been made (MIMIx: minimum information required for reporting a molecular interaction experiment) (49). Mechanisms for submitting annotations directly to the PPI databases in a unified format have also been developed (15).
Overall, our analysis lends strong support to the contention that curation policies play a key role in shaping the data collectively curated by PPI databases. These policies determine how useful the data are to the scientific community, in particular to the life scientists who routinely rely on these data for biomedical and clinical applications. Indeed, divergent organism assignments, the use of alternative protein identifiers, or different representation of complexes, although not reflecting actual curation errors, may lead to misinformation. These issues were raised in a recent study (28,30), which suggested that ‘errors’ of the type ‘wrong protein’ and ‘wrong organism’, among others, are not uncommon, and that the annotation of complexes as sets of spoke-expanded binary interactions is a potential source of concern. Our analysis has quantified these discrepancies on a global level, uncovering many more cases where pairwise discrepancies are attributable to similar issues (Examples 4–6 in the Supplementary Discussion S3).
Standardizing the curation policies along the lines advocated by the IMEx consortium, including the requirement for in-depth curation of articles (50), should go a long way towards resolving these issues. Members of the IMEx consortium also agreed to curate complementary sets of publications in order to increase coverage. We would like to suggest that this policy be revised to include a large enough number of commonly curated publications, in order to generate co-citations by IMEx members that can then be analyzed for compliance with the IMEx guidelines, using similar methods as those employed here.
The issues related to data curation are in no way limited to protein interaction data. Indeed, the importance of biological databases to the research community, combined with the rapid growth of the collected data, has highlighted a number of current limitations and needs related to the continued maintenance of such resources (51). Facing such challenges, some of the current efforts emphasize broader involvement of the research community in curation efforts (52), while others attempt to supplement or even replace manual curation with automated literature mining (53,54). But so far, the limitations of automated approaches only further underscore the many ambiguities and challenges of biocuration, indicating that manual curation is here to stay in the foreseeable future and that standardization of manual curation is an essential requirement.
Lastly, we examined the agreement level in co-citations of high-throughput articles and found it to be poor. This seems to be mainly due to the increased likelihood of differences occurring as the number of possible interacting entities grows, as well as to divergent policies for the annotations of large sets of raw versus filtered PPI data by each database (36). However, the small number of such articles contributes marginally to the discrepancies found in the co-citation the data set taken as a whole (Supplementary Discussion S2). Additional filtering of the data on the basis of various evidence codes, such as ‘interaction type’ or ‘interactions detection method’, was not performed mainly because the annotated information is frequently missing or too inconsistent to objectively evaluate agreement levels without a systematic re-examination of the original publications.
Each of the co-citations described in this article may be further explored using the ‘PubMed Report’ and ‘PubMed Detail’ utilities of the iRefWeb interface (http://wodaklab.org/iRefWeb/pubReport/), as described in detail in ref. (36). Further work with databases can now target those disagreements that are more likely due to genuine curation policy differences or curation errors. This is turn can lead to improved data curation policy and data that are more easily integrated, accessible and reliable. It is our hope that this study and its associated resources will contribute towards this goal.
Methods
Interaction data
Interaction data were consolidated from the following public databases and release dates, indicated in parenthesis: BIND (25 May 2005), BioGRID (7 September 2009), CORUM (8 September 2008), DIP (6 January 2009), HPRD (06 July 2009), IntAct (19 July 2009), MINT (28 July 2009), MPact (10 January 2008) and MPPI (6 January 2004). The consolidation was performed using the Interaction Reference Index process (18). iRefIndex examines amino acid sequences to establish the identity of proteins, instead of relying on gene or protein identifiers or database accession number, which are often subject to change. This enables it to reliably merge records from different databases that use distinct types of protein identifiers to support the same PPI.
The aggregated information comprised all the supporting evidence captured by the source databases using the PSI-MI controlled vocabulary (14). This includes the terms specifying the ‘interaction type’, the ‘interaction detection method’, and the corresponding literature citation, which is hyperlinked to the original PubMed identifiers. Discrepancies in the recorded information on the interaction type and detection method, while also revealing and important, were not globally monitored at this stage. The inherent ambiguities associated with this information make it very difficult to objectively quantify any detected discrepancies, let alone to interpret them.
Genetic interactions (39–41) were identified and marked for exclusion if their interaction types were defined as such by the source databases using the appropriate PSI-MI terms, as detailed in ref. (36). Inferred interactions from the OPHID database were likewise excluded (38). The iRefWeb resource (http://wodaklab.org/irefweb) provides details on the consolidation process, and views of the full original records as annotated by the source databases (36).
Quantifying the level of agreement
For all instances where two databases cite the same publication in their interaction record, we evaluate the agreement between the interactions and the proteins that they annotated from the publication. We denote such instances as ‘co-citations’. Depending on the number of databases citing the same publication, a single publication may give rise to several pairwise co-citations (Figure 1).
Sorensen–Dice similarity scores are non-normally distributed and display a different variance within different groups of co-citations. Therefore, the statistical significance of differences in SPPI and SProt distributions in distinct co-citation groups was computed using the non-parametric two-sample Kolmogorov–Smirnov test for equality of continuous distributions (implemented in R, http://www.r-project.org). Confidence intervals for the mean values of SPPI and SProt were computed using Student’s t-distribution, for groups containing at least 50 co-citations.
Mapping proteins to canonical isoforms and genes
Using our criteria, two databases would disagree on an interaction if they chose different peptide sequences to represent the same protein. Recording protein identifiers that point to different splice variants of the same gene is an important example of such discrepancies. Therefore, a further consolidation step was added to the iRefIndex procedure, whereby all the proteins were mapped to the canonical UniProt isoforms (56) of the corresponding genes, whenever possible [http://irefindex.uio.no/wiki/Canonicalization and ref. (36)]. This mapping was performed mainly to further normalize the consolidated data set. The level of agreement across the PPI landscape was measured both before and after isoform normalization.
Author Contribution
A.L.T. performed the systematic analysis of the consolidated data set of interactions. S.R. consolidated interaction data using the iRefIndex procedure. B.T. made the consolidated data amenable to analysis by implementing the iRefWeb database and query interface. I.M.D. supervised the iRefIndex project, performed initial analysis of the data, and provided illustrative test cases. S.J.W. supervised the iRefWeb project and the data-analysis project. A.L.T. and S.J.W. drafted the article.
Funding
Canadian Institutes of Health Research (MOP#82940); Canada Institute of Health Research (to S.J.W.); the Sickkids Foundation (to S.J.W.); Ontario Research Fund (to S.J.W.). Funding for open access charge: Canadian Institutes of Health Research (MOP#82940). S.J.W is Canada Research Chair, Tier 1.
Conflict of interest. None declared.
Acknowledgements
The authors wish to thank the teams of the BioGRID, DIP, HPRD, IntAct and MINT databases for their valuable comments on an earlier version of this article. Special acknowledgements are due to Sandra Orchard for helpful suggestions regarding iRefWeb features and to Gianni Cesareni for sharing a related manuscript prior to publication.



