-
PDF
- Split View
-
Views
-
Cite
Cite
Ankita Narang, Rishi Das Roy, Amit Chaurasia, Arijit Mukhopadhyay, Mitali Mukerji, Indian Genome Variation Consortium, Debasis Dash, IGVBrowser–a genomic variation resource from diverse Indian populations, Database, Volume 2010, 2010, baq022, https://doi.org/10.1093/database/baq022
Close - Share Icon Share
Abstract
The Indian Genome Variation Consortium (IGVC) project, an initiative of the Council for Scientific and Industrial Research, has been the first large-scale comprehensive study of the Indian population. One of the major aims of the project is to study and catalog the variations in nearly thousand candidate genes related to diseases and drug response for predictive marker discovery, founder identification and also to address questions related to ethnic diversity, migrations, extent and relatedness with other world population. The Phase I of the project aimed at providing a set of reference populations that would represent the entire genetic spectrum of India in terms of language, ethnicity and geography and Phase II in providing variation data on candidate genes and genome wide neutral markers on these reference set of populations. We report here development of the IGVBrowser that provides allele and genotype frequency data generated in the IGVC project. The database harbors 4229 SNPs from more than 900 candidate genes in contrasting Indian populations. Analysis shows that most of the markers are from genic regions. Further, a large fraction of genes are implicated in cardiovascular, metabolic, cancer and immune system-related diseases. Thus, the IGVC data provide a basal level variation data in Indian population to study genetic diseases and pharmacology. Additionally, it also houses data on ∼50 000 (Affy 50 K array) genome wide neutral markers in these reference populations. In IGVBrowser one can analyze and compare genomic variations in Indian population with those reported in HapMap along with annotation information from various primary data sources.
Database URL:http://igvbrowser.igib.res.in
Introduction
Indian population representing one-sixth of the world population has been the global melting pot of human diversity. It has all the world’s major linguistic groups and the populations have been shaped by different waves of migrations and admixture (1, 2). Further, stringent mating patterns have led to the existence of several endogamous populations, which makes it an important resource for mapping genes (3). The Indian Genome Variation Consortium (IGVC) project, an initiative of the Council for Scientific and Industrial Research (CSIR)—was set up to develop a database of genomic variations in Indian population for predictive marker discovery in complex diseases such as diabetes, asthma, neuropsychiatric, infectious and cardiovascular disorders, response to drugs, etc. (4). The Phase I of the project was conducted to determine the extent of genetic differentiation in India. Toward this genotype data of 405 SNPs from 75 genes and 4.2 Mb contiguous chromosome 22 regions were studied in 55 contrasting populations (4, 5). These populations were identified from 4 major linguistic groups namely, Austro-Asiatic (AA), Tibeto-Burman (TB), Indo-European (IE) and Dravidian(DR) spanning 6 geographical regions of habitat (N, north; NE, north-east; W, west; E, east; S, south; C, central) and different ethnic groups (LP, large population, caste; IP, isolated population, tribes; SP, special population, religious groups). Five genetically distinct clusters were identified and a set of 24 populations that represent these clusters were selected for the Phase II of the project. In the Phase II, 3824 SNPs from 834 candidate gene as well as ∼50 000 (Affy 50 K array) genome wide neutral markers have been genotyped using the illumina, sequenom and affymetrix platforms. This initiative lays the foundation for the integration of global genotype-to-phenotype data (6) with Indian population data and development of a federated database.
Data Source and Organization
To address the need for an online comprehensive resource that enables users to visualize IGVC data with integrated information about SNPs from different resources we have developed IGVBrowser as shown in Figure 1.
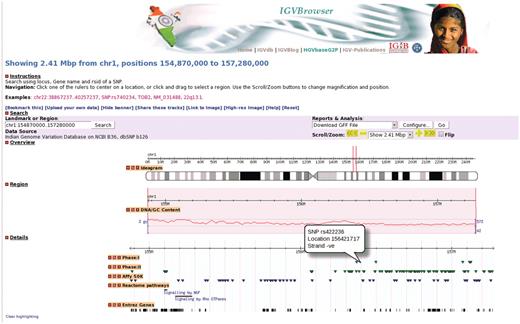
A representative example of IGVBrowser. Distribution of markers in 2.41 Mb region in human chromosome 1 from IGVC data is displayed along with annotation data from different resources.
IGVBrowser houses genotype data on samples that were recruited in the IGVC project. The database includes (i) final validated dataset from 1871 samples in Phase I comprising of 405 autosomal SNPs spanning over 75 genes including 90 SNPs from 5.2 Mb region of chromosome 22 from 55 diverse endogamous Indian populations (3); (ii) Phase II dataset for 3824 SNPs spanning from 834 genes in 545 samples from 24 IGVdb populations and (iii) ∼50 000 (Affy 50K XbaI array) neutral markers in 26 populations. The Phase II populations are a subset of the populations genotyped in the Phase I. Web-based tool SNPper (http://snpper.chip.org/) was used to classify the 4229 markers in Phase I and Phase II according to their location in genic regions (Figure 2). Similarly, DAVID (http://david.abcc.ncifcrf.gov/) was used to classify the genes containing these markers according to gene–disease association class (Figure 3) and their mapping in various KEGG pathways (Figure 4). We report that a large fraction of genes are implicated in cardiovascular, metabolic, cancer and immune system-related diseases. Thus, the IGVC data provide a basal level variation data in Indian population to study genetic diseases and pharmacology.
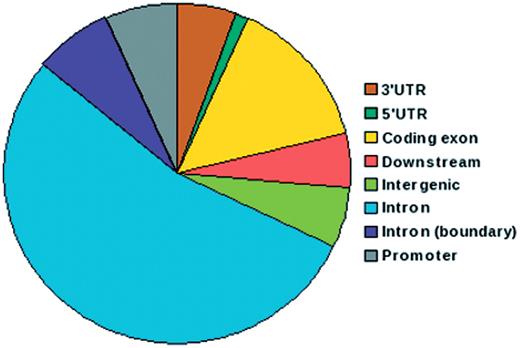
Pie chart depicting distribution of SNPs in IGVC according to genomic location. More than 50% of the SNPs belong to intronic regions and 15% are in coding exons.
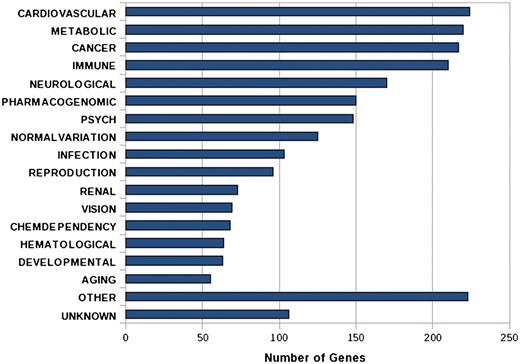
Bar graph shows the functional annotation of candidate genes in IGVC according to gene–disease association.
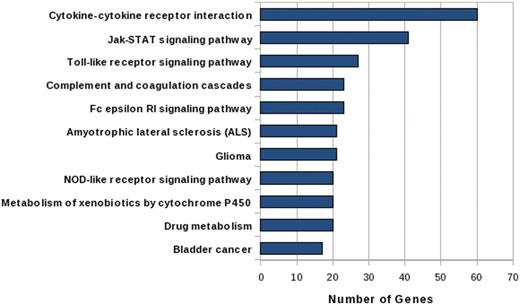
Bar graph shows the mapping of candidate genes in significant pathways (after Bonferroni correction) of KEGG Pathway Database.
IGVBrowser also included HapMap SNP genotype data from Phases I + II and III of the HapMap project (http://hapmap.ncbi.nlm.nih.gov/downloads/gbrowse/2009-02_phaseII+III/gff/) based on NCBI B36 assembly, dbSNP b126 from 4 populations: Yoruba from Ibadan, Nigeria (YRI); Japanese in Tokyo, Japan (JPT); Han Chinese in Beijing, China (CHB); and CEPH (Utah residents with ancestry from northern and western Europe) (CEU). Additional annotation information including cytogenetic positions, link to pathway annotations in the Reactome knowledgebase and mRNA sequences were retrieved from HapMap in Generic Feature Finding (GFF) format. Annotation data in tab-delimited format for non-coding RNA genes and pseudogenes, OMIM-associated Genes, miRBase and snoRNABase, simple repeats, database of genomic variants were downloaded from UCSC genome annotation database (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database) based on build hg18.
Database structure, implementation and accessibility
The browser implements one of the widely used platform-independent genome annotation viewer Generic Genome Browser (GBrowse v1.69), developed by Stein et al. (7) as a part of the Generic Model Organism System Database Project (http://www.gmod.org). GBrowse is a combination of database and interactive webpage for displaying genomic information along with providing data interoperability across systems running the same software. Integrated annotation data from primary sources like NCBI, UCSC and HapMap have been linked with variation data from different ethnic populations in India. Compiled data processed into GFF format and complete human genome sequence as plain text files were loaded into MySQL relational database management system using a script of GBrowse. IGVBrowser provides users an interactive display of the genetic variation data. A user can query chromosomal region of interest, reference SNP ID, HGNC symbols, pathway name or any other unique feature recognized by database as a query. It allows researchers to upload their own data in GFF format and view it along with data available in IGVBrowser. Semantic zooming feature of GBrowse in the IGVBrowser allows better interactive viewing options. In addition, the resource is facilitated with sequence analysis servers maintained by NCBI and UCSC. Online data analysis plugins allows text dumps of visible features using a number of standard formats and also facilitates the download of sequence corresponding to selected region.
Future directions
Indian Genome Variation data would be enormously useful for the dissection of common complex diseases and in pharmacogenomics studies. Frequency profiles of markers on disease or drug-related genes that have been generated through the IGVC are being used to identify at-risk chromosomes, founders, LD-based mapping, tracing history of diseases in pharmacogenetics as well as reference populations for mapping relatedness (3,4,5,8–19). The interactive web browser, IGVBrowser, has been created as a central repository for the current and future dataset on Indian populations and is being made accessible in the public domain. The web browser has been made dynamic for periodic future updates. A possible integration of IGVBrowser with HGVbaseG2P (20) can enable researchers for cross study comparison among different populations of the world for disease–gene association study.
Funding
Indian Genome Variation project was funded by the Council for Scientific and Industrial Research programme CMM0016 and SIP0006. Funding for IGVBrowser and open access charge is provided by European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 200754—the GEN2PHEN project.
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank Meenakshi Anurag, Pankaj Kumar for structuring the manuscript and Gajinder Pal Singh for correcting the draft and providing his valuable suggestions.



