-
PDF
- Split View
-
Views
-
Cite
Cite
Lucie N. Hutchins, Yueming Ding, Jin P. Szatkiewicz, Randy Von Smith, Hyuna Yang, Fernando Pardo-Manuel de Villena, Gary A. Churchill, Joel H. Graber, CGDSNPdb: a database resource for error-checked and imputed mouse SNPs, Database, Volume 2010, 2010, baq008, https://doi.org/10.1093/database/baq008
Close - Share Icon Share
Abstract
The Center for Genome Dynamics Single Nucleotide Polymorphism Database (CGDSNPdb) is an open-source value-added database with more than nine million mouse single nucleotide polymorphisms (SNPs), drawn from multiple sources, with genotypes assigned to multiple inbred strains of laboratory mice. All SNPs are checked for accuracy and annotated for properties specific to the SNP as well as those implied by changes to overlapping protein-coding genes. CGDSNPdb serves as the primary interface to two unique data sets, the ‘imputed genotype resource’ in which a Hidden Markov Model was used to assess local haplotypes and the most probable base assignment at several million genomic loci in tens of strains of mice, and the Affymetrix Mouse Diversity Genotyping Array, a high density microarray with over 600 000 SNPs and over 900 000 invariant genomic probes. CGDSNPdb is accessible online through either a web-based query tool or a MySQL public login.
Database URL:http://cgd.jax.org/cgdsnpdb/
Introduction
Single nucleotide polymorphisms (SNPs) are variable single base positions within a genome that represent the simplest and possibly most common type of genetic variation. Accordingly, SNPs have emerged as a powerful tool for tracking heredity and genetic variation, and have become especially popular for phenotype genome-wide association studies (1, 2). The critical role of the laboratory mouse has led to several efforts aimed at large-scale collection and analysis of mouse SNPs (3–7).
The Center for Genome Dynamics Single Nucleotide Polymorphism database (CGDSNPdb) was designed to bring together multiple sources of mouse SNP data, while checking them for accuracy and consistency among sources. CGDSNPdb is distinguished by the inclusion of two unique data sets:
The Imputed SNP Genotype Resource (IGR) (8) generated by a Hidden Markov Model (HMM) that assigns probable genotype and associated confidence levels for over 8 million SNPs in 74 strains of mice.
Data collected from over 140 strains of laboratory mice (filtered to 72 inbred strains in the current release, version 1.3) with the Mouse Diversity Genotyping Array [MusDiv; (9)], a high density microarray with probes that target 623 124 SNPs and over 900 000 invariant genomic regions targeting features such as exons and copy number variations. MusDiv SNP data will also be submitted to dbSNP following publication of an analysis manuscript (in preparation).
The CGDSNPdb search engine facilitates a number of different queries, including search by chromosome region(s), nearby gene annotations, or SNP identifiers. Results can be returned as dynamic html or in flat-text comma-separated-value (CSV) format.
Annotations in CGDSNPdb include characteristics of the SNP (e.g. presence in CpG dinucleotide, major/minor allele frequencies), along with functional characteristics of protein-coding genes affected by the SNP (e.g. changes in amino-acid physical and chemical characteristics, changes in codon usage, and overlapping or closest neighboring genes). All annotations were generated using an automated analysis pipeline with subsequent quality controls, described below.
CGDSNPdb was constructed primarily as a resource to support the imputation and mouse diversity array projects, however, it is being made available as a somewhat reduced size, but high confidence, collection of mouse SNPs. Database updates will be driven by the availability of new or updated genome assemblies, updated releases of major external SNP data sets, new SNP data sources, and maintenance. Future growth of the database will be targeted primarily at large-scale projects such as the ‘mouse genomes project’ (http://www.sanger.ac.uk/resources/mouse/genomes/) as well as data sets that can increase the represented strain diversity. Minor releases of CGDSNPdb may also be generated for improved data visualization or underlying quality control procedures. This manuscript provides a high-level overview of the main components of CGDSNPdb, as of version 1.3 (January 2010).
Implementation details
The database
CGDSNPdb was implemented using the open source MySQL relational database management system. The database consists of the core tables, containing all pertinent data for the SNP, including all data from the source download, and gene related tables that facilitate associations between SNPs and neighboring genes. Database schemas are available as Supplementary Figures S1 and S2. Original SNP files from sources and genome assembly files for flanking sequence data are retained and stored separately from the database.
Automated load programs, written in Perl and C++ with SQL queries, integrate data sets from various external sources into the database. These data files have, in general, been obtained directly from the generating source rather than another accumulative resource, such as dbSNP at NCBI (10) or the Mouse Phenome Database (MPD) (11). The load process (Supplementary Figure S3) includes a number of quality control checks that identify problems or ambiguities with SNPs and correct them, if possible. Quality control checks include comparison of the provided SNP call in C57BL/6J with the same position within the reference genome, genomic comparison of the provided flanking sequences (typically 50-nt up and downstream, with a requirement of at least 60% sequence identity in each direction), identification and resolution of duplicate entries (as defined by chromosome and position), and comparison of genotype calls (strain and genotype) among the different data sources. Genomic coordinates provided by the SNP source were assumed to be correct, and only challenged and further tested if the checks of the SNP or flanking sequence failed. No minimum length requirements were placed on the length of the flanking sequences, but SNP correction was only possible if flanking sequences were provided with the source data.
MusDiv (9) SNPs were subjected to additional quality tests intended to assist in the interpretation of the microarray hybridization patterns. The MusDiv SNPs hybridization probes are primarily 25-mers, with the SNP typically centered within the probe. Probes included both forward and reverse sense probes for both the reference C57BL6/J base and the known variant. Alignment of the flanking and probe sequences to the reference C57BL6/J genome was made using PASS (12), as it provided the best tradeoff of speed and alignment sensitivity (data not shown), especially for the analysis of near matches necessary for the mouse diversity array probes. PASS was used to align all four variants classes of probes, identifying all genomic matches with 0, 1 or 2 mismatches to the probe.
SNPs with conflicting genotype calls from different sources were loaded into the database, but flagged to indicate their conflict status. SNPs with conflicting duplicate calls from the same source were removed. SNPs with other unrecoverable errors (e.g. flanking sequences that do not align to the genome) were also flagged and removed as SNP entries in the database. SNPs in disagreement with the genome were identified and deleted prior to comparison for source-to-source conflicts, removing one systematic class of conflict. All details of the SNP processing and filtering are stored in log files that are subsequently loaded into the database, making deleted SNPs searchable, including the evidence that supports exclusion. Complete results of the CGDSNPdb data load processing and data quality check are available online at the CGDSNPdb website (http://cgd.jax.org/CGDSNPdbdb/utils/snp_data_report.php).
SNP-to-gene associations were generated through exhaustive testing of all SNPs in each data source with all transcripts currently in the ENSEMBL mouse (version 56_37i) transcript collection (13). SNPs that overlap genes with alternatively processed (splicing, transcription initiation, or polyadenylation) transcripts were compared against all isoforms in the collection. The comparison with all isoforms resulted in some SNPs with multiple classes of genomic location, since alternative processing can change whether or not the specific SNP is included in the final transcript (e.g. Figure 3).
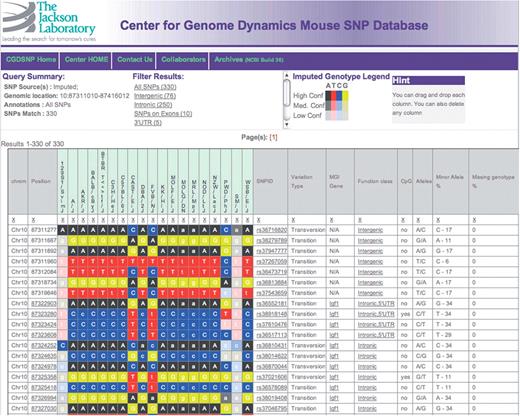
A screen capture of the standard summary page for SNP search, specifically showing results for the tumor suppressor P53, using the imputed data resource. Background colors in the genotype table represent the specific nucleotide and the imputed confidence level, with darkest colors representing the highest confidence.
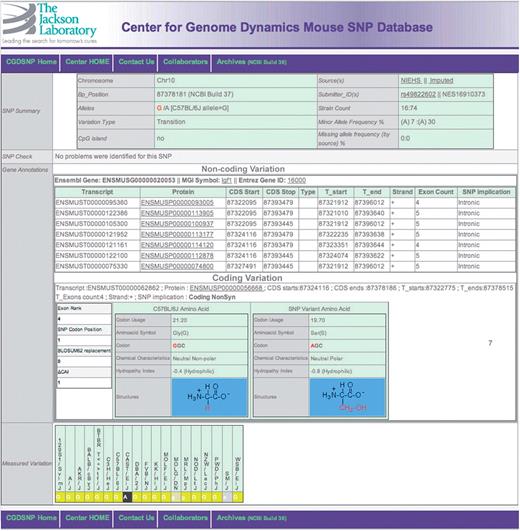
A screen capture for the SNP detail page, showing SNPs within the bounds of a transcript, whether in an UTR, intron or CDS. If multiple transcripts have been annotated for a given gene, the results are grouped by whether the change is within the coding sequence. A detail page for intergenic SNPs is available as Supplementary Figure S4.
CGDSNPdb currently (version 1.3) holds 9 686 537 distinct SNPs (Table 1), drawn from eight sources, including the previously mentioned IGR (8) and MusDiv (9) sets. Data were also drawn from large SNP collections generated by NIEHS (4), the Broad Institute (14), Genomics Institute of the Novartis Research Foundation (6), and Celera (15). Finally, two manually curated sets were included, specifically to support the needs of the IGR and MusDiv analysis. These included a set of SNPs targeted for studies of atherosclerosis and related diseases, drawn from a variety of sources (B. Paigen, personal comm.), and a collection of SNPs identified specifically for differentiating between wild-derived inbred and standard laboratory inbred strains, drawn from existing sources (10,11,16–18). A summary of the characteristics of all of our data sets is available here (Tables 2 and 3) and more detailed data are available at the CGDSNPdb web site.
| Classification . | Count . |
|---|---|
| Total | 9 686 537 |
| Transition | 6 607 155 |
| Transversion | 3 079 382 |
| Intergenic | 5 617 609 |
| Genic | 4 068 928 |
| Intronic | 3 850 229 |
| Exonic | 247 920 |
| UTR | 112 067 |
| CDS | 126 078 |
| CDS:nonsynonymous | 85 090 |
| CDS:synonymous | 43 698 |
| Noncoding gene exon | 17 032 |
| Noncoding gene intron | 5910 |
| Classification . | Count . |
|---|---|
| Total | 9 686 537 |
| Transition | 6 607 155 |
| Transversion | 3 079 382 |
| Intergenic | 5 617 609 |
| Genic | 4 068 928 |
| Intronic | 3 850 229 |
| Exonic | 247 920 |
| UTR | 112 067 |
| CDS | 126 078 |
| CDS:nonsynonymous | 85 090 |
| CDS:synonymous | 43 698 |
| Noncoding gene exon | 17 032 |
| Noncoding gene intron | 5910 |
| Classification . | Count . |
|---|---|
| Total | 9 686 537 |
| Transition | 6 607 155 |
| Transversion | 3 079 382 |
| Intergenic | 5 617 609 |
| Genic | 4 068 928 |
| Intronic | 3 850 229 |
| Exonic | 247 920 |
| UTR | 112 067 |
| CDS | 126 078 |
| CDS:nonsynonymous | 85 090 |
| CDS:synonymous | 43 698 |
| Noncoding gene exon | 17 032 |
| Noncoding gene intron | 5910 |
| Classification . | Count . |
|---|---|
| Total | 9 686 537 |
| Transition | 6 607 155 |
| Transversion | 3 079 382 |
| Intergenic | 5 617 609 |
| Genic | 4 068 928 |
| Intronic | 3 850 229 |
| Exonic | 247 920 |
| UTR | 112 067 |
| CDS | 126 078 |
| CDS:nonsynonymous | 85 090 |
| CDS:synonymous | 43 698 |
| Noncoding gene exon | 17 032 |
| Noncoding gene intron | 5910 |
| Source . | Available . | Loaded . | Strains . | Genomic mismatch . | Duplicate . |
|---|---|---|---|---|---|
| Imputed | 7 868 024 | 7 867 856 | 74 | 0 | 0 |
| MusDiv | 584 920 | 548 363 | 72 | 612 | 0 |
| NIEHS | 8 238 764 | 8 230 026 | 16 | 1830 | 22 620 |
| GNF | 156 513 | 155 677 | 76 | 611 | 243 |
| Broad | 138 602 | 138 594 | 48 | 233 | 9 |
| Celera | 2 122 060 | 2 122 059 | 5 | 0 | 0 |
| Paigen | 24 608 | 24 608 | 50 | 0 | 0 |
| Wild Derived | 667 | 667 | 37 | 0 | 0 |
| Source . | Available . | Loaded . | Strains . | Genomic mismatch . | Duplicate . |
|---|---|---|---|---|---|
| Imputed | 7 868 024 | 7 867 856 | 74 | 0 | 0 |
| MusDiv | 584 920 | 548 363 | 72 | 612 | 0 |
| NIEHS | 8 238 764 | 8 230 026 | 16 | 1830 | 22 620 |
| GNF | 156 513 | 155 677 | 76 | 611 | 243 |
| Broad | 138 602 | 138 594 | 48 | 233 | 9 |
| Celera | 2 122 060 | 2 122 059 | 5 | 0 | 0 |
| Paigen | 24 608 | 24 608 | 50 | 0 | 0 |
| Wild Derived | 667 | 667 | 37 | 0 | 0 |
| Source . | Available . | Loaded . | Strains . | Genomic mismatch . | Duplicate . |
|---|---|---|---|---|---|
| Imputed | 7 868 024 | 7 867 856 | 74 | 0 | 0 |
| MusDiv | 584 920 | 548 363 | 72 | 612 | 0 |
| NIEHS | 8 238 764 | 8 230 026 | 16 | 1830 | 22 620 |
| GNF | 156 513 | 155 677 | 76 | 611 | 243 |
| Broad | 138 602 | 138 594 | 48 | 233 | 9 |
| Celera | 2 122 060 | 2 122 059 | 5 | 0 | 0 |
| Paigen | 24 608 | 24 608 | 50 | 0 | 0 |
| Wild Derived | 667 | 667 | 37 | 0 | 0 |
| Source . | Available . | Loaded . | Strains . | Genomic mismatch . | Duplicate . |
|---|---|---|---|---|---|
| Imputed | 7 868 024 | 7 867 856 | 74 | 0 | 0 |
| MusDiv | 584 920 | 548 363 | 72 | 612 | 0 |
| NIEHS | 8 238 764 | 8 230 026 | 16 | 1830 | 22 620 |
| GNF | 156 513 | 155 677 | 76 | 611 | 243 |
| Broad | 138 602 | 138 594 | 48 | 233 | 9 |
| Celera | 2 122 060 | 2 122 059 | 5 | 0 | 0 |
| Paigen | 24 608 | 24 608 | 50 | 0 | 0 |
| Wild Derived | 667 | 667 | 37 | 0 | 0 |
| . | NIEHS . | MusDiv . | Broad . | GNF . | Celera . | Paigen . | Wild derived . |
|---|---|---|---|---|---|---|---|
| Imputed | 17 339 | 393 768 | 2793 | 5065 | 19 914 | 5374 | 19 |
| NIEHS | 37 916 | 5526 | 10 141 | 18 118 | 1048 | 0 | |
| MusDiv | 21 222 | 46 475 | 1343 | 2404 | 0 | ||
| Broad | 354 | 886 | 247 | 0 | |||
| GNF | 3292 | 6138 | 1 | ||||
| Celera | 728 | 1 | |||||
| Paigen | 0 |
| . | NIEHS . | MusDiv . | Broad . | GNF . | Celera . | Paigen . | Wild derived . |
|---|---|---|---|---|---|---|---|
| Imputed | 17 339 | 393 768 | 2793 | 5065 | 19 914 | 5374 | 19 |
| NIEHS | 37 916 | 5526 | 10 141 | 18 118 | 1048 | 0 | |
| MusDiv | 21 222 | 46 475 | 1343 | 2404 | 0 | ||
| Broad | 354 | 886 | 247 | 0 | |||
| GNF | 3292 | 6138 | 1 | ||||
| Celera | 728 | 1 | |||||
| Paigen | 0 |
Each cell counts for the number of SNPs where the indicated sources disagree on at least one strain genotype.
| . | NIEHS . | MusDiv . | Broad . | GNF . | Celera . | Paigen . | Wild derived . |
|---|---|---|---|---|---|---|---|
| Imputed | 17 339 | 393 768 | 2793 | 5065 | 19 914 | 5374 | 19 |
| NIEHS | 37 916 | 5526 | 10 141 | 18 118 | 1048 | 0 | |
| MusDiv | 21 222 | 46 475 | 1343 | 2404 | 0 | ||
| Broad | 354 | 886 | 247 | 0 | |||
| GNF | 3292 | 6138 | 1 | ||||
| Celera | 728 | 1 | |||||
| Paigen | 0 |
| . | NIEHS . | MusDiv . | Broad . | GNF . | Celera . | Paigen . | Wild derived . |
|---|---|---|---|---|---|---|---|
| Imputed | 17 339 | 393 768 | 2793 | 5065 | 19 914 | 5374 | 19 |
| NIEHS | 37 916 | 5526 | 10 141 | 18 118 | 1048 | 0 | |
| MusDiv | 21 222 | 46 475 | 1343 | 2404 | 0 | ||
| Broad | 354 | 886 | 247 | 0 | |||
| GNF | 3292 | 6138 | 1 | ||||
| Celera | 728 | 1 | |||||
| Paigen | 0 |
Each cell counts for the number of SNPs where the indicated sources disagree on at least one strain genotype.
Web interface
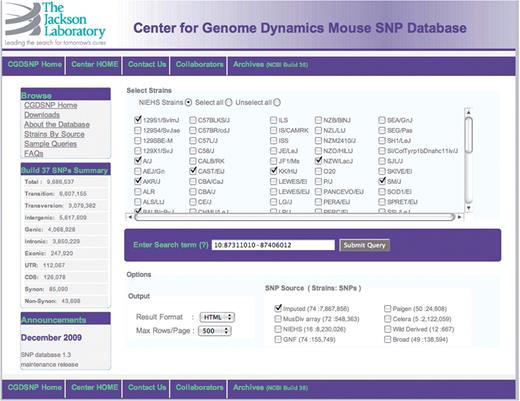
The web interface to CGDSNPdb was built using open source Linux-Apache-MySQL-PHP (LAMP) tools. The database is searchable through an online interface (Figure 1) that can accept as input genome coordinates, SNP accession ids, ENSEMBL gene predictions and Mouse Genome Informatics (MGI) gene symbols (11) and Entrez gene IDs (19). Searches can be restricted to source of SNP data, and specific strains of interest. As noted above, choices of data source include the IGR and MusDiv sets (Figure 1). Pop-ups are used for help tips such as the acceptable formats for a search term and the data sources that include SNPs for any particular strain of mouse.
All searches return a common result page (Figure 2), which includes a summary of the parameters of the search followed by a table representation of the SNP genotypes and annotations. Currently displayed annotations include SNP accession ids (from all sources that match a SNP), mutation type (transition or transversion), MGI gene symbol (11), ENSEMBL transcript (13), CpG site status, minor allele frequency and frequency of failed SNP assay. The returned table is a dynamic object that allows the user to re-order or remove individual columns. The summary page also contains links that allow further restriction of the displayed SNPs by class of location within the gene annotations on the genome, including intergenic, exonic, intronic, etc. All displayed data from external sources such as MGI or ENSEMBL, as well as genomic location within the CGD genome browser, are active links that enable direct access to the appropriate external websites. As of version 1.3, ENSEMBL annotations are for mouse release 56_37i, and MGI annotations were downloaded as of 12 January 2010.
In the HTML representation, the result table contains both text and background color representations of the genotype data for the selected strains (Figure 2). When the imputed SNP genotypes are included in the analysis, the background color intensity is varied to show the ‘confidence level’ associated with the imputed call, with darkest colors representing high confidence and lightest colors representing low confidence (Figure 2). Experimentally assessed and imputed genotypes are shown as upper and lower case letters, respectively. Genotypes with a conflict between data sources are shown as question marks if both sources are included in the query.
Every SNP is also further described in a SNP detail page (Figure 3). The SNP detail page takes on different forms depending upon the genomic nature of the SNP location. All SNP detail pages display a standard set of information that includes: data source and source-specific identification, genomic location (chromosome and base position), major and minor alleles, a summary of the results of quality control analysis, and the class of SNP location. Further data display on the SNP detail page is dependent on whether or not the specific SNP is located within the genomic boundaries of any annotated transcripts (Figure 3).
The detail page for SNPs within the genomic bounds of a transcript, whether within coding sequence (CDS), untranslated region (UTR), or introns includes details of the gene and transcript(s) that were matched, including reference genomic coordinates, with links to external databases. CDS SNP descriptions also include the rank of the exon within the transcript structure, position within amino-acid codon, and functional characteristics of the variants, including both codons and their usage frequencies in mouse, both amino acids and their associated BLOSUM62 substitution score (20), side chain chemical characteristics, hydropathy indexes, and the molecular structure of each amino acid (Figure 3). The detail page for Intergenic SNPs (Supplementary Figure 4) includes information on the immediate neighboring genes, including relative orientation and distance to the SNP.
Access
The CGDSNPdb webserver interface URL is http://cgd.jax.org/cgdsnpdb/. CGDSNPdb can also be accessed directly with read-only privileges at host cgd.jax.org. Login instructions are available on the webserver interface. Raw data files, including those for the MusDiv and IGR can be found at http://cgd.jax.org/datasets/popgen.shtml.
Additional features
Additional features available at the online site include user help pages, a Frequently Asked Questions (FAQ) page, and database report pages. The database report page provides details generated automatically during the process of preparing and inserting the external data sources into the database, including numbers of SNPs processed, number inserted successfully, and numbers with each of the types of problems and/or inconsistencies. Users can ultimately drill down to obtain lists of SNPs that fall into each specific category.
Future Plans and Upgrades
Future plans include increased integration of additional annotation data that assess the functional implications of the variation, including but not limited to intersection of SNP locations with known or predicted functional elements such as splice sites, promoter elements, and polyadenylation sites or overlap with regions of evolutionary conservation, such as provided by the PhastCons scores (21). The extended ambiguity analysis implemented for the MusDiv (9) SNPs and associated array probes will be extended to all SNPs in the database for which flanking sequence is available. Finally, search capabilities will be improved with further integration of gene identification data, such as Unigene (22) IDs. Any additional upgrades will be focused on requests generated by users of the database.
Citing CGDSNPdb
The CGDSNPdb should be referenced using this publication. In addition, since CGDSNPdb is planned to include regular updates, references to use of the database should include the current database version and month and year of access.
Funding
National Centers for Systems Biology program grant GM-076468 from the National Institute of General Medical Sciences. Funding for open access charge: National Institute of General Medical Sciences.
Acknowledgements
The authors thank Carol Bult and three anonymous referees for critical review of the manuscript.
Conflict of interest. None declared.



