-
PDF
- Split View
-
Views
-
Cite
Cite
Reza M. Salek, Kenneth Haug, Pablo Conesa, Janna Hastings, Mark Williams, Tejasvi Mahendraker, Eamonn Maguire, Alejandra N. González-Beltrán, Philippe Rocca-Serra, Susanna-Assunta Sansone, Christoph Steinbeck, The MetaboLights repository: curation challenges in metabolomics, Database, Volume 2013, 2013, bat029, https://doi.org/10.1093/database/bat029
Close - Share Icon Share
Abstract
MetaboLights is the first general-purpose open-access curated repository for metabolomic studies, their raw experimental data and associated metadata, maintained by one of the major open-access data providers in molecular biology. Increases in the number of depositions, number of samples per study and the file size of data submitted to MetaboLights present a challenge for the objective of ensuring high-quality and standardized data in the context of diverse metabolomic workflows and data representations. Here, we describe the MetaboLights curation pipeline, its challenges and its practical application in quality control of complex data depositions.
Database URL:http://www.ebi.ac.uk/metabolights
Introduction
Metabolomics is an emerging research field, which provides a snapshot of the metabolic dynamics that reflect healthy metabolome or response of living systems to pathophysiological stimuli and/or genetic modification. Metabolomics is a fast-growing discipline, with numbers of publication in peer-reviewed journals rising steadily every year. Similarly to other ‘-omics’, there is a great need to share and disseminate metabolomics data, making data accessible to the public, as funding organizations and journals increasingly require it. Therefore, MetaboLights was set up as a medium to capture metabolomics base investigation. The MetaboLights repository was officially launched on 28 June 2012 at the 8th International Conference of the Metabolomics Society in Washington DC, USA (1, 2). MetaboLights to date already incorporates ∼160 metabolomics-related protocols and ∼1000 assays, which span over nine different species including human, Caenorhabditis elegans, Mus musculus and Arabidopsis thaliana. Currently, nearly 1600 metabolites have been identified in these studies and mapped to different databases, from which ∼1000 have been mapped to Chemical Entities of Biological Interest (ChEBI) (3). These studies cover a variety of techniques, including nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry. An extensive set of associated information for studies is stored and displayed in MetaboLights. This includes submitter and author information, publication references, the study design, protocols applied, names of data files included, platform information and metabolite information. The metabolite information includes a description, external database identifiers, formula, simplified molecular-input line-entry system (SMILES), The IUPAC International Chemical Identifier (InChI) and intensity or concentration, and where the metabolite was identified in the sample. Depending on the technology we also capture identified metabolite-relevant information such as chemical shift and multiplicity for NMR-based experiments, and m/z, retention index, fragmentation and charge for mass spectrometry.
MetaboLights data are free to download and use for any purpose as per the standard EMBL-European Bioinformatics Institute (EMBL-EBI) terms and conditions (http://www.ebi.ac.uk/Information/termsofuse.html). All public studies are downloadable, as individual files or packaged as full zip file generated on demand in ISA-Tab metadata file format (4) including the associated instrumental data files, directly from the online study details page, and from the MetaboLights download page http://www.ebi.ac.uk/metabolights/download. A direct bulk download using ftp is available from ftp://ftp.ebi.ac.uk/pub/databases/metabolights/, which is organized into sub-folders for public studies. An online search facility provides the ability to explore content using free text through a standard ‘lucene search engine’, indexing the underlying data fields, including the study description, study title, protocols, metabolites and authors. Currently, we support free-text searching and users can combine multiple search terms, for example, ‘human urine’ will give you all studies where you find the terms ‘human’ and ‘urine’ are used. Users can browse a complete list of all publicly available studies in MetaboLights, and if the user is registered and currently logged into MetaboLights, additional private studies may be displayed. These private studies are either under the user’s control or have been directly shared from other users. It is possible to further refine the search result using ‘facets’. Search and browse facets give users the ability to limit the search/browse results to a selection of species, platform and metabolites.
We have seen a demand from a growing number of publishers and public funding agencies for greater transparency, with data sets and the results from studies being made publicly available through submission to repositories. This makes it possible for the study to be accessible for examination by the wider metabolomics community. Data sets can be used as a knowledge or education resource and provide means for collaboration initiatives across different groups and fields. As more data sets become available, MetaboLights will also become an invaluable resource for bioinformaticians to develop new algorithms or tools for processing of metabolomic data. MetaboLights allows laboratories across the globe to collaborate on metabolomics projects through data sharing, and thereby to begin to generate collaboratively the large data sets needed to address how environmental or dietary factors can modulate the metabolome. The use of the ISA framework, adopted by the growing ISA Commons (5) community will also ensure a certain level of interoperability with an increasingly diverse set of life science domains and other data types.
We have introduced MetaboLights to the metabolomics community with several earlier publications (1, 2) as well as presentations at the relevant scientific meetings. We also actively promote MetaboLights via several social media sites, including the following:
Twitter—@metabolights
Facebook—http://www.facebook.com/metabolights
We have also carried out workshops introducing data submission steps and usage of the MetaboLights website via the EMBL-EBI Industry Programme and MetaboLights Project Workshop as well as a Cambridge University Course: Bioinformatics: Metabolomics Data and Tools, introducing MetaboLights and usage of ISA tools in capturing experimental metadata. http://ruddles.bio.cam.ac.uk/∼dpjudge/Descriptions/Metabolomics.php.
MetaboLights is a registered bioDBcore (6) resource in the BioSharing catalogue (http://www.biosharing.org/biodbcore) and in the MIRIAM registry (7) (http://www.ebi.ac.uk/miriam).
The present contribution does not include content previously reported on, such as the user features that the MetaboLights database provides (1) or a description of the standards coordination initiative COSMOS (2). Rather, this contribution focuses specifically on the curation pipeline for metabolomics content in MetaboLights, and the challenges, which we are experiencing in that regard. The remainder of this article is organized as follows. The next section describes the submission pipeline and the challenges, which we have experienced in optimizing standardization of content in the challengingly diverse field of metabolomics. The section thereafter recounts post-submission curation efforts and techniques that we have implemented. Finally, we outline some future developments and give our concluding remarks.
The MetaboLights submission pipeline
The first step in getting quality data in a curated repository is to gather as much metadata as possible in a standardized format at the time of submission. Submissions to MetaboLights rely on the ISA-Tab format as a vehicle for experimental metadata and data files (Figure 1). The input file in ISA-Tab format can be created and edited using two main recommended routes. The first is the ISAcreator software application: a standalone, Java-based, platform independent desktop application with a range of facilities to enable standards-compliant creation of ISA-Tab archives. The software enables ontology searches and lookups with a great deal of flexibility for capturing metadata at various stages of the experimental workflow. These include sample preparation and extraction protocols, instrument-related parameters and related metadata, all of which comply with the ‘Metabolomics Standards Initiative’ (MSI) reporting recommendations (8, 9).
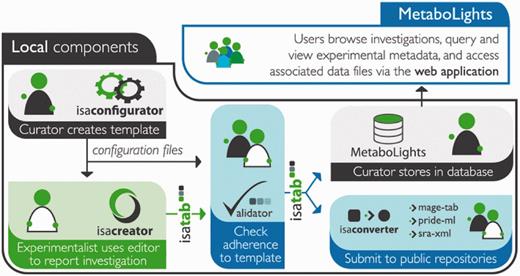
Showing a typical submissions pipeline using the ISA suite and submission to MetaboLights.
For users who do not wish to install a standalone application in their environment, the second route available for submissions is via the use of MetaboLights Google templates, which combine the ISA-Tab syntax, Google spreadsheets and the functionality of the ‘OntoMaton’ widget (10) (http://isatools.wordpress.com/2012/07/13/introducing-ontomaton-ontology-search-tagging-for-google-spreadsheets/). OntoMaton is a Bioportal-powered (11) add-on to Google Spreadsheets that brings semantic support for the use of ontologies to create standardized annotations and metadata. The result is ISAcreator-like features available within the Google collaborative environment, a handy feature for all groups dealing with multi-user multi-centre studies. The comparison of the two different ISA-Tab metadata capturing routes is summarized in Table 1.
Comparison of the two main recommended route to capture experimental metadata
| Submission route . | Domain . | Automated annotation . | Ontology search/lookup . | Versioninga . | Collaboration . |
|---|---|---|---|---|---|
| ISAcreator | Multiomics | ✓ | ✓ | ✗ | ✗ |
| OntoMaton | General | ✓ | ✓ | ✓ | ✓ |
| Submission route . | Domain . | Automated annotation . | Ontology search/lookup . | Versioninga . | Collaboration . |
|---|---|---|---|---|---|
| ISAcreator | Multiomics | ✓ | ✓ | ✗ | ✗ |
| OntoMaton | General | ✓ | ✓ | ✓ | ✓ |
aBy versioning we refer to managing of user edits throughout the annotation process.
Comparison of the two main recommended route to capture experimental metadata
| Submission route . | Domain . | Automated annotation . | Ontology search/lookup . | Versioninga . | Collaboration . |
|---|---|---|---|---|---|
| ISAcreator | Multiomics | ✓ | ✓ | ✗ | ✗ |
| OntoMaton | General | ✓ | ✓ | ✓ | ✓ |
| Submission route . | Domain . | Automated annotation . | Ontology search/lookup . | Versioninga . | Collaboration . |
|---|---|---|---|---|---|
| ISAcreator | Multiomics | ✓ | ✓ | ✗ | ✗ |
| OntoMaton | General | ✓ | ✓ | ✓ | ✓ |
aBy versioning we refer to managing of user edits throughout the annotation process.
However, creating ISA archives for submission into MetaboLights is not confined to these two access routes. In fact, any tool able to create ISA-Tab documents may be used to submit to MetaboLights. This facility is broadly accessible because there are a broad range of libraries supporting the ISA-Tab syntax. (For example, Perl, Python, Java and R libraries supporting the syntax are available from the ISA-tools pages on GitHub, https://github.com/ISA-tools). Two resources [Golm database (12) and MetabolomeXpress (13)] are currently setting up direct pipelines of this sort: one, Golm, is creating ‘push submissions’ using its own exporter, while MetabolomeXpress is facilitating ‘pull submissions’ in that conversion to ISA-Tab is performed by MetaboLights from local metadata files. These facilities will be extended to foster collaborations with other major metabolomic resources to allow data exchange and replication between all key metabolomics nodes. When it comes to actual data files as opposed to the associated metadata, MetaboLights is pragmatic: all of ‘raw’ instrumental data formats, converted open-source file formats and any format of processed data are supported. However, we strongly recommend that processed data should be made available in open formats, ideally alongside the analysis workflow (e.g. R or MATLAB routines) that was used to generate those outputs.
MetaboLights-specific configuration of ISA-Tab
Work is currently being undertaken to refine the annotation requirements for the submission pipeline and to provide canonical gas chromatography–mass spectrometry, liquid chromatography–mass spectrometry and 1D NMR metabolomic experiment representations to guide submitters and developers alike. ISAconfigurations, the configuration files used by the ISAcreator toolkit, for these techniques will be issued, detailing annotation parameters for metabolomic specific configurations, including vocabulary support (via ontology lookup) within each specific ISA-Tab section. Complementing these coding guidelines, work is underway to refine the description of data matrix semantics to ensure better practices for reporting experimental findings and conclusions.
Plugins added by MetaboLights
To report metabolite findings and identification events, an additional ‘Metabolite Identification plugin’ has been added to ISAcreator (Figure 2). Based on the OSGi plugin architecture, it allows capture of key information for all small molecules identified in a study, with a link to a relevant chemical database. We considered using PubChem (14) the most comprehensive source for metabolite identification. Because PubChem comprises several different chemical databases, we use the open Web Services (programmatically accessible search) to retrieve relevant chemical information such as Chemical Formula, Compound Names, SMILES and InChI, once the correct metabolites have been chosen from the suggested list by the submitter (Figure 2). The link to the identified metabolites could be from any of the relevant chemical database entries, such as ChEBI, HMDB (15) or Lipid Maps (16). Various database entries, where possible, would be replaced by ChEBI identifier by the curation team, more details to follow. The plugin is compatible with mzTab (https://code.google.com/p/mztab/), a tab-delimited file format originally designed to capture proteomics data having the objective of allowing biologists to be able to open the files in Excel and be able to ‘see’ the data. Recently, an R package has been developed to facilitate data analysis. The ‘Risa module’, available since BioConductor release 2.11 (http://www.bioconductor.org/packages/2.12/bioc/html/Risa.html), includes functionality to process mass spectrometry data relying on the XCMS package (17), and provides methods to save analysis results back to ISA archives, therefore allowing data provenance tracking at the source, while reducing the annotation burden on scientists.
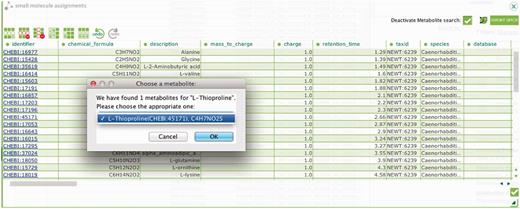
The ISAcreator plugin, capturing metabolites identified within the metabolomics experiments and mapped to the relevant chemical database.
Data submission challenges
In response to challenges with the speed of response in our submission pipeline, we have implemented a new queueing system for online submission, as well as an ftp service for bulk uploads of larger data sets. The submission queue allows the user to upload an experiment and then continue browsing while MetaboLights is validating, storing and indexing the experiment. The queue system is available for new submissions, updates and ‘public release date’ changes. However, there was a remaining issue in that large data files (with sizes well exceeding 100 GB) still gave problems for our pipeline. To address this, we have followed the direction taken by other ‘-omics’ and gene-sequencing databases in relying on ‘portable hard drives’ and FTP transfer protocols. We are also working collectively with PRIDE (18) to implement Aspera-based next-generation high-speed file transfers, and a standalone package that can work silently in the background to upload bulk data sets.
Challenges in data quality, metabolite identification and curation
Metabolomics is a relatively new member of the ‘-omics’ field, with instrument-related technologies rapidly changing and evolving, becoming more accurate and more sensitive at the same time. Metabolomics also has the most diverse range of instruments used to capture the biological metabolome matrix for a diverse range of biological samples in comparison to other -omics, with each technology requiring a wide range of parameters to be controlled and reported. Due to the diverse nature of metabolomes from different biological sources, the sensitivity, characterization and limitation of technology platforms used to capture this data, the existence of various methods for samples preparation, extraction and modification, the application of numerous column ranges for chromatography and separation technologies and the existence of various methods for data processing and handling reproducibility of metabolomics studies is known to be limited and challenging (19). Aspiring to address this challenge, MetaboLights requests data submitters to provide all protocols used for sample manipulation using free text, and to provide important parameters separately using controlled terms from applicable ontologies within ISAcreator. We try to have a balance between the relevant and important information captured and the time required to complete the task by adhering to MSI (5). One objective is to abstract various protocols into controlled sets of standard operating procedures available for re-use by submitters, facilitating experimental reproducibility and making similar instrumental data sets more comparable. In addition, where possible, we capture not only the actual instrument raw output files, for each of the samples, but also the quality controls (QC), replicated samples (technical or biological), blank samples and reference chemical compounds used for metabolite identification (Figure 3). Metabolite identification is a hotly debated issue within the metabolomics community. The reporting requirement and evidence needed to accurately and reliably identify a metabolite is time-consuming to adhere to, requiring additional experimental work, additional data acquisition time and producing larger file sizes, including data such as ms/ms, msn and 2D NMR (19, 20). Since March 2012, all metabolites originating from data submitted to MetaboLights is being curated into the ChEBI database. This ensures we have more control over metabolite identification and that high-quality curated metabolites information is available to the community. These curated metabolites are associated with experimental data in the repository, enabling links between chemical structures, raw metabolomics data and biological context to be derived. Metabolites submitted to the MetaboLights repository are then first fully checked and matched (via the ChEBI ontology) with existing metabolites entire in ChEBI. If an equivalent was missing from ChEBI, that metabolite would be curated in ChEBI by their curation team and eventually reported back to MetaboLights to supplement or replace the existing ones.
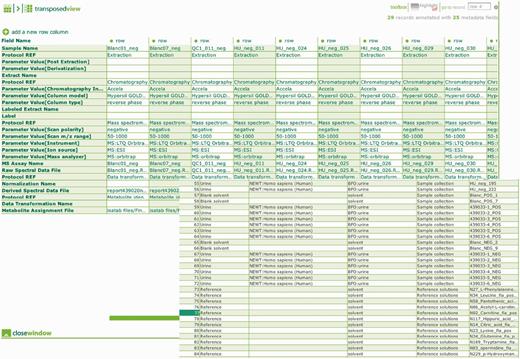
Overview of the ontology link instrumental and experimental metadata captured within ISAcreator, example shows metadata for the samples, QC, blanks and reference chemical compounds.
Once a metabolomics study has been uploaded to MetaboLights and passed the ISA-Tab structure validation process, a unique MetaboLights identifier is assigned to the study. This stable (permanent) accession number is used to uniquely identify, and is the online access point, for the processed study information. At this point, our curation team will be notified about the new submission, which then would be examined by the curators, verifying whether correct information has been captured using the ISA-tool structure and whether it adheres to the MSI and correctly links to the relevant ontology. Currently, this is a manual process and requires constant communication between the curator and submitter for clarification and corrections of the parameters and factors within the study. Some studies fail during the validation process, notifying the MetaboLights team automatically. We will then contact the submitter to assist with the submission process. This involvement is sometimes even sooner when dealing with complex experimental settings and usage of various analytical instruments and assays; at this point, curation works start before any submission to MetaboLights, working with the user towards a complete and curated submission.
Enhancing the data collection and curation future plans
Our future curation plan once metabolite entries are curated in ChEBI, is then to assign the identified metabolites to pathways in Reactome (21). In addition to further help address data quality challenges, development of a curation tool that will allow expert curators or reviewers to examine and suggest corrections to experimental data submitted to MetaboLights is currently underway. We are planning to add a new section with ‘Gold Standard Studies’ that could potentially be used for easy reference consisting of known good quality experiments, which includes replicates, QC samples and reference compounds. These data sets can then be used as templates for examining experimental factors, and be used for data mining or as a reference for the development of bioinformatics tools and novel data processing techniques. We are developing several new components to enhance the data collection and curation process. These include a Glyph-based graphical rendering for experimental workflows and ISA2OWL/ISA2RDF conversion modules. These will be released in early 2013. The former will provide a unique ability to provide a ‘bird’s eye view’ of the experimental design, while the latter will enable connecting to linked data clouds and also enable more advanced data validation. It is expected that both new modules will further support data curation and improve the standard in experimental data representation.
Future plans for a curated reference layer
The MetaboLights repository is being extended with a reference layer/knowledge base of curated quality data about metabolites (1), scheduled to launch in Summer 2013. ‘The Reference Layer’ will be based around a curated metabolite-centric view and will include comprehensive knowledge elements such as reference spectra of various types, biological reference data, protocols, cross-references to other resources and advanced search and download functionality. This metabolomics offering would be manually curated and will integrate with existing EMBL-EBI services, for example including chemical structures and characteristics from ChEBI (3), metabolic pathways from Reactome, and other external sources such as HMDB (15) for reference spectroscopy and chromatography spectra, reference biology, occurrence and concentration in species, organs, tissues and cellular compartments in various conditions. We will also share publication references and protocols.
We recognize that good user-centred design is key for the success of a web service and database (22). To best achieve this, we organized a workshop in April 2012 at Hinxton together with a dedicated EBI-user experience expert to help us with the design of the MetaboLights Reference Layer curation pipeline and tools. Several users with diverse metabolomics backgrounds were recruited to participate in various exercises, like a metabolite mind-mapping session, giving the opportunity to design metabolite-centric workflows. The feedback was then used in the implementation process starting with low-fidelity mock-ups and eventually progressing to a full web interface and portal for the reference layer. Currently, we have prototyped the main search functionality for the reference layer and the compound-centric view, linking this information to pathways, reactions, organisms and sources, literature and spectra arising from both of the main technologies (NMR and MS).
There are no web services for programmatic access available at present. However, this functionality is scheduled for a future release of the repository.
Future of metabolomics standards in COSMOS
An EU coordination action for developing metabolomics standards in the EU and worldwide, called COordination of Standards in MetabOlomicS—COSMOS (http://cosmos-fp7.eu), was launched in October 2012. The MetaboLights team is coordinating this consortium of 14 European partners, with MetaboLights playing a central role for the proposed work. A key aspect of this effort aims to develop efficient policies ensuring that metabolomics data are encoded in open standards, tagged with a community-agreed and complete set of metadata, supported by a communally developed set of open-source data management and capturing tools disseminated in open-access databases adhering to these standards, supported by vendors and publishers, who require deposition on publication, and properly interfaced with data in other biomedical and life science e-infrastructures. Our aim is to deliver the exchange formats and terminological artefacts needed to describe, exchange and query metabolomics experiments, using the ISA-Tab as core for the description of experiments and building additional ‘layers’ for the data matrices. We wish to ensure that the proposed standards are widely accepted by involving major global players in the development process. We will also develop and maintain exchange formats for raw data and processed information (identification, quantification), building on experience from standards development within the Proteomics Standards Initiative (23). Additionally, we are planning to collaborate on developing the missing open standard NMR Markup Language for capturing and disseminating NMR spectroscopy data in metabolomics. We aim to explore semantic web standards that facilitate linked open data throughout the biomedical and life science realms, and demonstrate their use for metabolomics data.
Open-access source code and documentation
The MetaboLights Repository source code is regularly updated and is publicly available at http://sourceforge.net/projects/metabolomes, and include details on how to install a local version of MetaboLights. The ‘ISAcreator Metabolite Identification Plugin’ can be found at: https://github.com/EBI-Metabolights/ISAcreatorPlugins.
To facilitate user feedback, we have created a SourceForge tracker for logging issues, available at http://sourceforge.net/projects/metabolomes.
There is also an online contact form, http://www.ebi.ac.uk/metabolights/contact, and a contact email address, metabolights-help@ebi.ac.uk as well as comprehensive online help to guide the data submission process, which includes online video instructions. The submission guide is available at http://www.ebi.ac.uk/metabolights/submitHelp.
Conclusion
We have described the submission and curation pipelines of the MetaboLights repository for metabolomics, and highlighted pressing challenges together with on-going efforts to improve the curation utilities surrounding the database offering. MetaboLights is in its infancy compared with other well-established ‘-omics’ databases such as Pride, ArrayExpress and others, but already acts as an accretion point. We anticipate it will become a cornerstone resource in the same way these proteomics and transcriptomics repositories are in terms of breadth and depth. However, there is a need to have control over the quality and complexity of the metabolomics data sets right from the beginning. The complexity and variety of metabolomics technologies makes a fully automated data deposition system challenging. In addition, curators have to work closely with the data submitters to be able to understand different workflows and experimental settings. There are wide varieties of proprietary file formats by different instrument vendors in use, with challenges such as lack of transparency, interoperability and data sharing, as well as potential costs of licensing. Hence, there is a need and requirement of usage and promotion of open-source file formats that we hope to facilitate and promote via our COSMOS consortium and with the help of the metabolomics community for acceptance and usage. Once open-source formats are accepted and in use, it is possible to automate capturing instrumental metadata using relevant tools and parsers, as long as the formats are also supported by software producers and instrument vendors. There is a need to develop a set of robust validation procedures that can identify and highlight missing or potentially erroneous elements of metabolomics experiments using an ‘inspector tool’, not only to visualize the raw files but to ensure the quality of publicly available metabolomics data. All of these developments synergize to promote and further improve the reproducibility and transparency of research involving data generation and exchange in the field of metabolomics.
Acknowledgements
The MetaboLights project team would like to thank the following persons for their invaluable contributions (names are alphabetically ordered): Rafael Alcántara, Masanori Arita, Mike Beale, Nick Bond, Kees van Bochove, Jildau Bouwman, Steve Bryant, Hong Cao, Juan Castrillo, Jenny Cham, Cecilia Castro, Tim Ebbels, Michael Eiden, Oliver Fiehn, Andrew Gibbs, Roy Goodacre, Martin Hornshaw, Jan Hummel, Albert Koulman, Peter Meadows, Pablo Moreno, Theo Reijmers, Francis Rowland, Linda Scoriels, Mark Seymour, Tim Smith, Anthony Taylor, Chris Taylor, Michael Wakelam, Jane Ward and David Wishart.
Funding
The development of MetaboLights is funded by the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/I000933/1 to C.S]. Funding for open access charge: [BBSRC BB/1000933/1]. The ISA framework is supported by the BBSRC [grants BB/I025840/1, BB/I000771/1 and BB/J020265/1] and the University of Oxford e-Research Centre (funding to S.A.S.). COSMOS is funded by European Commission [grant EC312941 to C.S. and S.A.S.].
Conflict of interest. None declared.
References
Author notes
Citation details: Salek,R.M., Haug,K., Conesa,P., et al. The MetaboLights repository: curation challenges in metabolomics. Database (2013) Vol. 2013: article ID bat029; doi: 10.1093/database/bat029



