-
PDF
- Split View
-
Views
-
Cite
Cite
Pushp Priya, Mukesh Jain, RiceSRTFDB: A database of rice transcription factors containing comprehensive expression, cis-regulatory element and mutant information to facilitate gene function analysis, Database, Volume 2013, 2013, bat027, https://doi.org/10.1093/database/bat027
Close - Share Icon Share
Abstract
Rice is one of the most important crop plants, representing the staple food for more than half the world’s population. However, its productivity is challenged by various stresses, including drought and salinity. Transcription factors (TFs) represent a regulatory component of the genome and are the most important targets for engineering stress tolerance. Here, we constructed a database, RiceSRTFDB, which provides comprehensive expression information for rice TFs during drought and salinity stress conditions and various stages of development. This information will be useful to identify the target TF(s) involved in stress response at a particular stage of development. The curated information for cis-regulatory elements present in their promoters has also been provided, which will be important to study the binding proteins. In addition, we have provided the available mutants and their phenotype information for rice TFs. All these information have been integrated in the database to facilitate the selection of target TFs of interest for functional analysis. This database aims to accelerate functional genomics research of rice TFs and understand the regulatory mechanisms underlying abiotic stress responses.
Database URL:http://www.nipgr.res.in/RiceSRTFDB.html
Introduction
Substantially larger proportion of world's population depends on rice for food requirement. However, exposure to abiotic stresses results in enormous losses in rice production. Among various abiotic stresses, drought and salinity are the major concern, as they are the major factors responsible for heavy yield losses. At the molecular level, these stresses affect the expression of several genes, referred as stress-responsive genes (1, 2). A large number of stress-responsive genes have been identified in various plants, including rice (3–7). Although many of these genes have been functionally analyzed, the exact molecular mechanism(s) underlying various abiotic stress responses is still unknown. In addition, there is a great need to identify most suitable target genes for engineering stress tolerance in crop plants like rice.
Plant breeding and genetic engineering approaches are generally adopted to impart or enhance stress tolerance in plants. Genetic engineering methods involve development of transgenics by modulating one or more potential key regulators of transcriptional circuits to enhance stress tolerance and understand the complex mechanism behind stress responses. Among several stress-responsive genes, transcription factors (TFs) are the most suitable targets to unravel the molecular mechanisms of abiotic stress responses and engineering abiotic stress tolerance in plants, as they can act as master regulators controlling the expression of many target genes (8–11). However, the role of only a few rice TFs in abiotic stress responses have been elucidated until now and require detailed investigations.
The web-accessible database providing information on various genomic elements in different plant species proves to be an invaluable resource for researchers involved in crop improvement. A few attempts have been made to develop databases, which provide information on stress-responsive genes in plants. A Plant Stress Gene Database based on literature search and another database of annotated tentative orthologs from crop abiotic stress transcripts are available (12, 13). In addition, a few instances of species-specific stress-related databases are also available. Stress-responsive TranscrIption Factor DataBase (STIFDB) provides information related to Arabidopsis genes up-regulated during abiotic stress together with putative TF-binding sites (14). Recently, the information about abiotic stress-related quantitative trait loci (QTLs) has been integrated with rice genomic data in QliCRice (a web interface for abiotic stress-responsive QTL and loci interaction channels in rice) database (15). A few other databases providing various omics information for rice, including Rice TOGO Browser (http://agri-trait.dna.affrc.go.jp/) for integrated information on functional and applied genomics, OryzaExpress (http://bioinf.mind.meiji.ac.jp/OryzaExpress/) for gene expression networks and omics annotation and Rice Tos17 Mutant Panel Database (http://tos.nias.affrc.go.jp/) for information on rice mutants, are also available (16–19). However, a dedicated database for rice TFs containing related information, which could provide a platform for their large-scale functional analysis in stress responses, is still lacking.
In this study, we designed a web-accessible database, RiceSRTFDB (Rice Stress-responsive Transcription Factor Database), which in addition to the expression profiles under stress conditions and various tissues/developmental stages, provides access to cis-regulatory elements and mutant information for rice TFs. The information provided in the database in combination with advanced experimental approaches may provide a foundation for analyzing the function of individual TF to help understand the regulatory mechanisms involved in abiotic stress responses.
Methods and database contents
Transcription factors in rice
A total of 2478 non-redundant set of rice TFs was compiled from Plant Transcription Factor Database (PlnTFDB, http://plntfdb.bio.uni-potsdam.de/v3.0/) and Database of Rice Transcription Factors (http://drtf.cbi.pku.edu.cn/) (20, 21). These TFs were classified into 84 families, including one orphan family, based on their domain composition following the rules for each family given in PlnTFDB. The largest number of TFs belong to AP2-EREBP family (165) followed by bHLH (160) and NAC (144) families. The orphan family contains 58 TFs, which could not be classified in any other TF family. The genomic information about all the TF encoding gene loci, including locus ID, chromosome position, gene description, gene coding sequence and protein lengths and number of exons and introns was extracted from the Rice Genome Annotation Project (RGAP) database (http://rice.plantbiology.msu.edu/; version 6.0) (22). The corresponding Rice Annotation Project Database (RAP-DB, http://rapdb.dna.affrc.go.jp/) (23) gene locus IDs along with hyperlink have also been provided in the database. In addition, we identified the presence of conserved domain(s) in each TF using InterproScan. The Arabidopsis best hit for each rice TF has also been identified by BLAST search for comparative analysis. All these information for each rice TF have been integrated into RiceSRTFDB.
Stress-responsive gene expression
The availability of a large volume of microarray data provides an opportunity to study the gene expression at whole-genome, gene family or single gene level and identify the gene(s) involved in various biological processes (24). Genome-wide transcriptome analyses of various rice tissues under drought and salinity stress conditions using microarrays have been carried out earlier (25–28) and data is available in the public databases. To study the expression profiles of rice TFs, we used microarray data available at the Gene Expression Omnibus (GEO) database generated using Affymetrix platform because it shows high degree of reproducibility and homogeneity across different laboratories when compared with other platforms. For this, we first identified the probesets present on the Affymetrix microarray chip corresponding to each TF encoding gene loci. Of 2478, no probeset was found for 202 TFs and among the remaining, 1647 showed unique (one-to-one) mapping to the probeset. Other 629 TFs showed multiple (many probesets mapped to single locus) or ambiguous (single probeset mapped to multiple loci) mappings. About 85% of the multiple/ambiguous mappings were resolved by manual curation and assigned a unique probeset representing 3′-end of the TF gene loci. However, the ambiguous mapping of probesets to 93 TFs could not be resolved and thus were not included in gene expression analysis.
Microarray data from a curated set of 456 Affymetrix GeneChip Rice Genome arrays, including 99 arrays representing 18 different drought and salinity stress treatments and 357 arrays representing 131 tissues/organs/developmental stages, was used (Supplementary Table S1). Microarray data was normalized using RMA algorithm implemented in GeneSpring software (v11.0). We used data from 18 different drought and salinity stress treatment conditions to identify stress-responsive TF genes (Supplementary Table S1). The genes showing change of at least 2-fold at a P-value of ≤0.05 after ANOVA analysis under at least one stress condition analyzed were identified as differentially expressed TFs. A total of 1408 (56.8%) TFs were found to be differentially expressed under at least one stress condition analyzed. Out of 84, at least one member of 76 TF families showed differential expression (Supplementary Table S2). The largest number of AP2-EREBP family members exhibited differential expression followed by bHLH, MYB and NAC families (Figure 1). A significant number (29) of TFs included in orphan family also showed response to drought and salinity stress conditions. At least 10 members of 38 TF families were differentially expressed under stress conditions. Among the 48 families containing >10 members, more than half the members of 38 families exhibited altered gene expression under stress conditions. Interestingly, >80% members of Aux/IAA, C2C2-CO like and Tify families showed differential expression in response to drought and salinity stress conditions. The differential expression of several members of these TF families under stress conditions has previously also been reported (6, 7, 29–31).
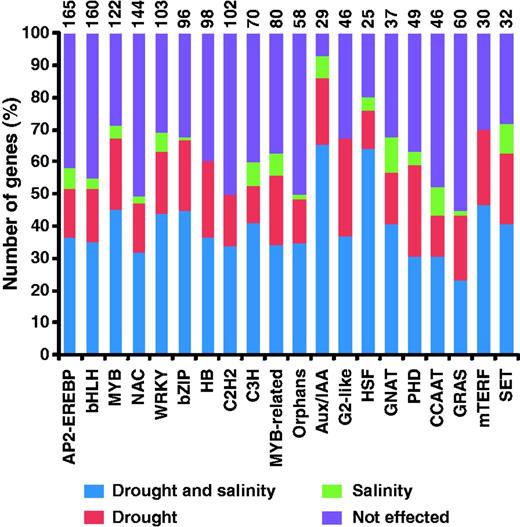
Top 20 families of rice TFs with largest number of genes showing differential expression under drought and/or salinity stress conditions. ‘Not effected’ represents genes showing no significant change in expression under drought and/or salinity stress conditions. The total number of TFs included in each family is given at the top of each bar.
Further, we identified TF encoding genes showing overlapping and specific responses to drought and/or salinity stress conditions (Figure 2A). The accessibility to these set of genes along with their response under stress condition(s) has been implemented in RiceSRTFDB and links to various related information provided. Among the 1308 and 938 TFs differentially expressed under drought and salinity stresses, respectively, 838 are commonly up- or down-regulated under both conditions. Another 470 and 100 TF genes showed specific response to drought and salinity stress, respectively. Some of the commonly regulated TFs exhibited opposite response under drought and salinity stresses. We found that stress response of a few TFs specifically regulated under drought or salinity stress was genotype-, tissue- and growth stage-specific. These observations suggest that same TF can have different roles under different stress conditions and/or tissues/development stages. Further, we studied the stress response of TFs in specific tissues used in microarray analysis. Under drought stress, a larger number of TF genes showed down-regulation in 7-day-old seedling and roots from different stages of development (Figure 2B). However, the number of up-regulated TF genes was higher in leaves from different developmental stages under drought stress. Overall, the largest number of TF genes was regulated in seedling followed by reproductive stages of development (leaves and root at panicle elongation stage and young panicle). A larger number of TFs were found to be regulated in 10-day-old seedlings as compared with roots under salinity stress (Figure 2C). Previous reports also showed the higher sensitivity of rice plants to different stresses at seedling and panicle development stages (6, 7, 32).
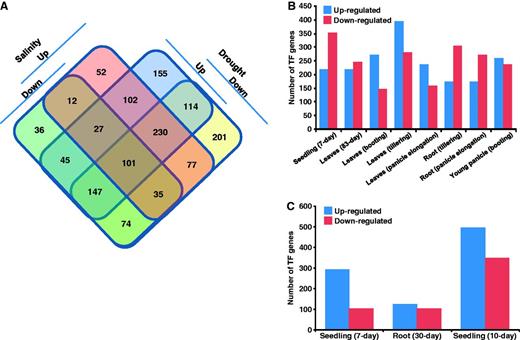
Stress-responsive gene expression of rice TFs. (A) Venn diagram showing the number of genes up- and/or down-regulated under drought and/or salinity stress conditions. (B) Number of TF genes up- and down-regulated in different tissues/developmental stages under drought stress. (C) Number of TF genes up- and down-regulated in different tissues under salinity stress.
Further, to study the gene expression of rice TFs during development, we have integrated their expression in various tissues/organs and developmental stages as well. It will facilitate the identification of target TF genes expressed in a particular tissue/developmental stage and involved in stress response. Many evidences have demonstrated the interaction of developmental processes and stress responses (3, 5, 29, 33).
Tos17 mutant information
The analysis of mutants with disrupted gene sequence is the most efficient strategy to study gene function. To facilitate the functional analysis of rice genes, a collection of ∼50 000 Tos17 insertion lines has been generated and phenotyped comprehensively (16, 34). The phenotypic data of all the lines along with Tos17 flanking sequences are available in the Rice Tos17 Insertion Mutant database (http://tos.nias.affrc.go.jp/). These mutants have been used for functional analysis of several rice genes (35–38). The identification of putative Tos17 mutants for rice TFs and their phenotypes would accelerate their functional analysis. Therefore, we investigated the availability of mutants for rice TFs in the Tos17 mutant panel database. To identify potential insertion of Tos17 that might affect the regulatory activity of rice TFs, 92 589 Tos17 flanking tag sequences (downloaded from NCBI) were searched against both genomic and 2 kb upstream sequences of TFs using BLAST. The flanking tags showing at least 90% similarity with 90% coverage were considered to be significant and assigned to the corresponding TF. The flanking tags located in the genomic and promoter region for 519 TFs (403 in genomic region and 194 in promoter region) could be identified. Among these, 3070 mutant lines could be identified corresponding to 451 TFs. The phenotype data were available for 1723 mutant lines representing 356 TFs, which have been integrated in the RiceSRTFDB. Among the various observed phenotypes, low fertility was most frequent, followed by dwarfism in the mutant lines showing insertion in genomic or promoter regions of TFs (Supplementary Figure S1). Several mutant lines showing alterations in reproductive organs and developmental stages (tillering, heading, panicle, flower and seed) were also identified. From these data, it can be speculated that rice TFs play a crucial role in overall plant growth and might be responsible for tissue-/developmental stage-specific stress responses.
cis-regulatory elements
Fundamentally, the regulation of gene expression is mediated by the binding of TFs to specific cis-regulatory elements present in the promoter sequence of their target genes. One or more TF(s) can regulate the expression of other TF(s) by binding to their cis-regulatory elements, which result in complex regulatory networks for highly controlled and specific pattern of gene expression. A few such transcriptional regulatory networks operative in response to abiotic stresses have been defined in Arabidopsis and rice (8). Therefore, the identification of cis-regulatory elements is important to elucidate the molecular mechanism of abiotic stress responses. To reveal the presence of various cis-regulatory elements in the promoter, 2 kb upstream region of each rice TF gene was analyzed by PLACE database (http://www.dna.affrc.go.jp/PLACE/index.html) search. Based on the available knowledge in the literature, the identified cis-regulatory elements were categorized as stress-responsive and other motifs in RiceSRTFDB by manual curation. A variety of stress-responsive elements present in the promoters of rice TFs were identified. Among these, MYB-binding motif, dehydration-responsive element and abscisic acid-responsive element were most frequent. The integration of expression data and cis-regulatory elements along with experimental data will provide insights into the regulatory interactions among TFs and other proteins regulating stress responses in rice.
Web-accessible RiceSRTFDB design
To make the data publicly available, all the data/results were stored in MySQL relational database. A diagrammatic representation of data integrated in the RiceSRTFDB is given in Supplementary Figure S2. The snapshots of various utilities and output format are given in Figure 3. The web interface for Rice SRTFDB was developed using Perl CGI and Javascript running on the Apache web server. The graphs used to represent expression patterns were generated using Perl module GD graph.
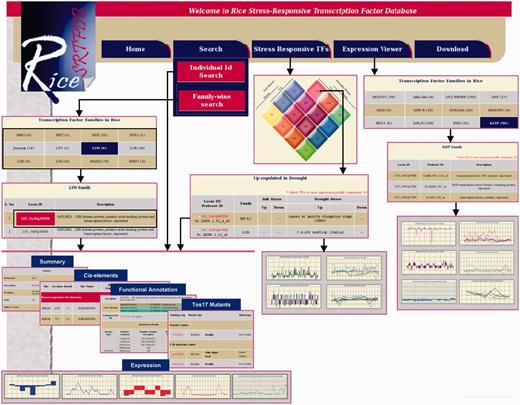
Snapshots of the RiceSRTFDB showing various search functions and result output.
The database can be queried either TF family-wise or using RGAP gene locus IDs. Distinct sections have been created to present comprehensive information about each TF, which includes summary, functional annotation, Tos17 mutants, stress-responsive cis-elements in the promoter region and expression under drought and salinity stresses and at various developmental stages. Hyperlinks to RGAP, RAP-DB (for corresponding RAP gene loci), TAIR (for Arabidopsis best hit) and NCBI (for flanking tag sequence of Tos17 insertion) have also been provided to facilitate access to sequences associated with each rice TF. Based on their response to various salinity and drought conditions, different classes of TFs have been represented as interactive Venn diagram with clickable links, which displays the list of TFs included under respective class along with their expression in different stresses and developmental stages/tissues. Expression viewer facilitates the visualization of expression patterns of TFs belonging to particular family during different stress and specific developmental stages. Besides, all the data included in database have also been provided for download for large-scale analysis.
Conclusions and future direction
In this post genomics era, the most important challenge is to analyze and present the increasing size of data in a meaningful way to make it usable for all the researchers. RiceSRTFDB is a user-friendly web interface, which provides a range of information about the rice TFs for public access. This database will decrease the need for curation of various genomic information about rice TFs by researchers. The availability of integrated comprehensive information, including expression profiles, cis-regulatory elements and mutant(s) availability for individual or family-wise TF(s) together in the database is expected to prioritize the functional analysis of TFs of interest. We believe that the data/information integrated in the database will support basic and applied research to understand the complex regulatory mechanism of stress responses and engineering stress tolerance. The database will be updated on regular basis with the availability of new gene expression data and availability of mutants. Further, with an increasing amount of data, additional information related to the rice TFs, including expression patterns in different cultivars and genomic variations will also be integrated in the database in future. We intend to incorporate gene expression data on other abiotic stresses as well. Additional features may also be included in the database to cater the demands of researchers.
Funding
This work was financially supported by the Science and Engineering Research Board (grant number SR/S0/PS/07/2011), Department of Science and Technology, Government of India and core grant from NIPGR. Funding for open access charges: NIPGR.
Conflict of interest. None declared.
References
Author notes
Citation details: Priya, P. and Jain, M. RiceSRTFDB: a database of rice transcription factors containing comprehensive expression, cis-regulatory element and mutant information to facilitate gene function analysis. Database (2013) Vol. 2013: article ID bat027; doi:10.1093/database/bat027



