-
PDF
- Split View
-
Views
-
Cite
Cite
Kalpana Raja, Suresh Subramani, Jeyakumar Natarajan, PPInterFinder—a mining tool for extracting causal relations on human proteins from literature, Database, Volume 2013, 2013, bas052, https://doi.org/10.1093/database/bas052
Close - Share Icon Share
Abstract
One of the most common and challenging problem in biomedical text mining is to mine protein–protein interactions (PPIs) from MEDLINE abstracts and full-text research articles because PPIs play a major role in understanding the various biological processes and the impact of proteins in diseases. We implemented, PPInterFinder—a web-based text mining tool to extract human PPIs from biomedical literature. PPInterFinder uses relation keyword co-occurrences with protein names to extract information on PPIs from MEDLINE abstracts and consists of three phases. First, it identifies the relation keyword using a parser with Tregex and a relation keyword dictionary. Next, it automatically identifies the candidate PPI pairs with a set of rules related to PPI recognition. Finally, it extracts the relations by matching the sentence with a set of 11 specific patterns based on the syntactic nature of PPI pair. We find that PPInterFinder is capable of predicting PPIs with the accuracy of 66.05% on AIMED corpus and outperforms most of the existing systems.
Database URL:http://www.biomining-bu.in/ppinterfinder/
Introduction
Protein–protein interactions (PPIs) are of central importance to understand the mechanisms of biological processes and diseases (1). The knowledge about PPIs is rapidly growing with the results from high-throughput experimental technologies. Accordingly, a huge number of interaction data are being published in the literature (1, 2). A wide range of interaction databases such as IntAct (3), MINT (4), BIND (5) and DIP (6) have been developed by manually curating the protein interactions from various information sources. However, the rapid growth of biological publications in recent years made this time-consuming task almost impractical for the PPI extraction. Consequently, many of the PPI data are still available only in the literature (7). Extraction of such information from biomedical literature has become an important topic in the field of biomedical natural language processing (BioNLP) (8).
Several approaches ranging from simple co-occurrence principle to advanced NLP and pattern matching techniques (9) to more sophisticated machine learning methods (10) have been reported for extracting PPI information. Among them, NLP techniques are most popular and highly preferred for PPI extraction for more than a decade. These techniques can be referred as parsing methods with the possibility of shallow and full parsing to produce the output as constituent trees or dependency trees (11–13). However, full parsing yields potentially better results than shallow parsing because of its elaborate syntactic information. Extraction of PPI information from the parsed sentences depends on the syntactic pattern of two proteins and the relation keyword. These patterns are sequence of words, or part-of-speech tags describing the relation between two proteins in a biomedical text. Pattern matching technique looks for a match in a sentence with at least two proteins and a relation keyword (14).
In general, extraction of PPI from literature broadly consists of two components, protein name recognition and PPI extraction, both of which are equally challenging. Though many approaches have been proposed for the extraction of PPI information from the biomedical literature, the problem still remains as an open challenge for the researchers to develop more accurate, robust and automated methods to address the problem. One alternative to this challenge is to develop PPI systems specific to one organism (e.g. human) and such systems are very few (15). In this article, we present PPInterFinder, a PPI extraction tool for mining human PPIs from the biomedical literature. The tool integrates NLP techniques (Tregex for relation keyword matching), rule sets (comprise of seven rules) for identifying candidate PPI pairs and finally pattern matching algorithm (three abstract forms and 11 extended patterns) for PPI relation sentence extraction. This unique mining tool specific to humans is helpful to the users to find and extract both known and potentially novel human PPIs from literature.
Materials and methods
Architecture and components
PPInterFinder is a web-based tool for mining human PPIs. The project is a combination of Java libraries for relation keyword recognition, negation recognition, candidate PPI pair identification, pattern matching and PPI information extraction. In addition, a Perl module implementing Perl/CGI scripts is used for web interface designing to upload user input data. The work flow of the system is as follows: (i) text preprocessing; (ii) candidate PPI pair recognition and PPI information extraction. Figure 1 shows a general work flow of the system.
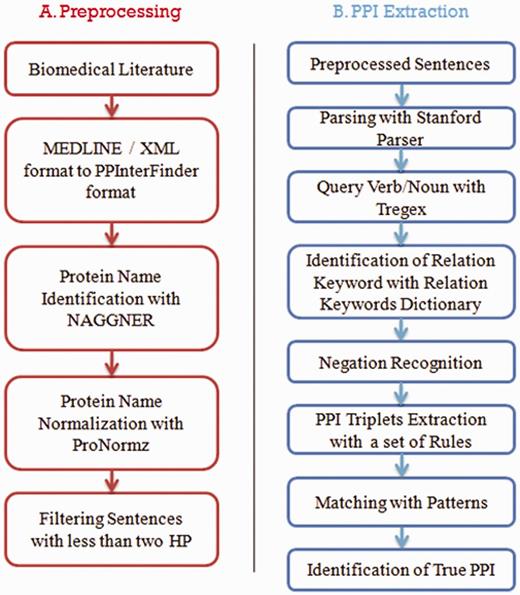
Text preprocessing
The input text can be a PubMed abstract in plain text format or MEDLINE/XML format with unique PubMed ID. An initial preprocessing is carried out to match PubMed IDs with individual sentences in the abstract. Further processing includes (i) identification and normalization of protein names and (ii) filtering out of input sentences with only one protein or no protein names. The protein name recognition and normalization are carried out by our own tools, namely, NAGGNER (16) and ProNormz (17), which are highly specific to human proteins.
Extraction of PPI information
Relation keywords dictionary
The success of PPI system relies on the successful identification of relation keyword. To achieve this goal, we have developed a vast relation keywords dictionary, which consists of 354 relation keywords. The keywords are grouped into 88 subtypes by identifying the common root word for each subgroup (Supplementary Data 1). The relation keywords dictionary is created on the basis of various keywords used in the previous articles related to PPI extraction (9, 18–20) and further augmented with relation keywords from other interaction databases such as IntAct (3), MINT (4) and DIP (6).
Relation keyword recognition
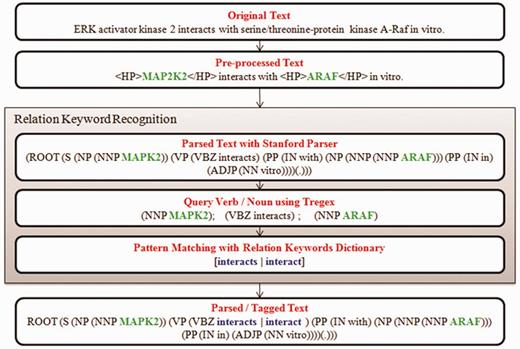
The relation keyword can either be a verb or a noun and its recognition is a vital step prior to the extraction of PPI information. The text mining and NLP methods implemented in the identification of relation keyword are illustrated in Figure 2. First, the input sentence is parsed using Stanford Parser (21) with grammar settings to englishPCFG module to generate the constituent tree of verb and noun phrases. Next, the node labels of verb/noun are queried using a tree query language called Tregex (22), a Java API developed within the Stanford Parser package for querying expressions of a parse tree. Tregex expressions are very similar to regex expressions (java.util.regex library), but more advanced. Finally, the algorithm performs a pattern matching between verb/noun words against the relation keyword dictionary and the final matching word is declared as the relation keyword.
Negation keyword recognition
The success of every automated PPI extraction from biomedical literature invariably depends on the proper recognition of negation keywords (10). Most of the available PPI extraction systems consider the negation keyword, ‘not’ to avoid false PPI extraction (9, 10). In the present study, we consider the recognition of three keywords ‘no’, ‘not’ and ‘neither/nor’ as negation keywords as these keywords are mostly associated with false PPI information in human PPIs sentences. These negation keywords normally occur as an adverb (e.g. ‘not’), a determiner (e.g. ‘no’) or a coordinating conjunction (e.g. ‘neither/nor’). The algorithm locates the presence of any negation keyword in the parsed sentence through pattern matching, similar to relation keyword recognition.
Abstract forms for PPI candidate pair
In biomedical text, the relationship between two entities (protein–protein) can be expressed in different abstract forms (18, 23). We use the following three types of ‘abstract forms’ depending on the position of the relation keyword co-occurring with two proteins.
| Form 1: | PROTEIN1 - token* - RELATION - token* - PROTEIN2 |
| Examples: | PROTEIN1 interacts with PROTEIN2 |
| PROTEIN1 has weak association with PROTEIN2 | |
| Form 2: | RELATION - token* - PROTEIN1 - token* - PROTEIN2 |
| Example: | interaction between PROTEIN1 and PROTEIN2 |
| Form 3: | PROTEIN1 - token* - PROTEIN2 - token* - RELATION |
| Example: | PROTEIN1 and PROTEIN2 complex |
| Form 1: | PROTEIN1 - token* - RELATION - token* - PROTEIN2 |
| Examples: | PROTEIN1 interacts with PROTEIN2 |
| PROTEIN1 has weak association with PROTEIN2 | |
| Form 2: | RELATION - token* - PROTEIN1 - token* - PROTEIN2 |
| Example: | interaction between PROTEIN1 and PROTEIN2 |
| Form 3: | PROTEIN1 - token* - PROTEIN2 - token* - RELATION |
| Example: | PROTEIN1 and PROTEIN2 complex |
Form 1 is the most common form with relation keyword in between a pair of proteins (protein–relation–protein). In Form 1, the relation keyword is commonly a verb, verb with additional tokens/words or even a noun. Form 2 and Form 3 are comparatively rare with relation keyword at the corners (relation–protein–protein or protein–protein–relation). In such cases the relation keyword is mostly a noun.
Rule set for identification of candidate PPI pairs
We incorporate seven rules for extracting candidate PPI pairs from sentences related to the three abstract forms discussed above (Table 1). The various forms of our seven rules to extract candidate PPI pairs include (i) the position of relation keyword with a pair of proteins (Rule 1), (ii) the number of tokens/words between the protein pairs (Rule 2), (iii) simple sentences with two proteins (Rule 3), (iv) simple sentences with two proteins and a negation keyword (Rule 4), (v) complex sentences having more than two proteins (Rule 5), (vi) complex sentences having more than two proteins and a negation keyword (Rule 6) and (vi) complex sentences having three proteins and two negation keywords (Rule 7). All the seven rules and their role in extracting true PPI pairs are explained below.
Rule 1 is mandatory to understand the position of relation keyword with a pair of proteins. The relation keyword may appear either between the proteins (protein–relation–protein) or at the corners (relation–protein–protein or protein–protein–relation) as described in three abstract forms earlier. Furthermore, the relation keyword will be commonly a verb or noun in Form 1 and will be a noun in Forms 2 and 3. This grammatical information of the relation keyword helps in eliminating many false PPIs. For example, if the relation keyword matched is not verb or noun in abstract Form 1, then it is considered as false PPI.
The number of tokens/words between the entities (two proteins and a relation keyword) varies widely in all abstract forms. However, the number of tokens/words between the proteins in abstract Forms 2 and 3 is very important to avoid false PPI extraction. Rule 2 confirms the presence of one token between the protein pair in abstract Form 2 and one or no token between the protein pair in abstract Form 3.
PPI extraction procedure is simple for sentences with two proteins and a relation keyword matching the abstract Form 1. An additional step is required for candidate PPI pairs in sentences matching the abstract Forms 2 and 3. In such cases, Rule 3 looks for the number of tokens/words between the protein pair as per Rule 2. Examples 1 and 2 illustrate the extraction of PPI information from sentences in abstract Forms 1 and 2, respectively.
PubMed ID: 11909642: <PROTEIN> MAP2K2 </PROTEIN> <RELATION> interacts </RELATION> with </PROTEIN> ARAF <PROTEIN>in vitro.
PubMed ID: 15208391: The <RELATION> association </RELATION> between <PROTEIN> CAND1</PROTEIN> and <PROTEIN> CUL1 </PROTEIN> - TAP is specific.
The approach is very similar to Rule 3, except the role of negation keyword to filter false PPI information. Example 3 illustrates the importance of negation keyword in the recognition of non-interacting protein pairs.
PubMed ID: 16899217: There was <NEGATION> no </NEGATION> detectable <RELATION> interaction </RELAION> between <PROTEIN> PSMC6 </PROTEIN> and <PROTEIN> PSMC5 </PROTEIN>.
We use an algorithm for Rule 5 as illustrated in Figure 3. The complexity of the algorithm depends on the number of proteins present in the input sentence.
The word position is assigned to each word in the sentence, starting from 0.
A hash table is generated to hold proteins, relation keyword and their corresponding word position.
The relation keyword in the hash table is identified.
All possible PPI triplets are generated by combining the relation keyword with each of the preceding and succeeding proteins.
Finally, all the true PPIs are declared.
The algorithm is very similar to Rule 5 with an additional check for the presence of negation keyword to avoid false PPI extraction. The proteins following the negation keyword are considered to be false PPIs and subsequently eliminated.
Rule 7 is explicit for sentences having the negative keyword ‘neither/nor’. We observed that such sentences comprise a minimum of three proteins and a relation keyword. The false PPIs are identified by the specific order of the entities as shown in Example 4.
PubMed ID: 12007405: <NEGATION> Neither </NEGATION> <PROTEIN> SLCO6A1 </PROTEIN> <NEGATION> nor </NEGATION> <PROTEIN> BRI1 </PROTEIN> <RELATION> interact </RELATION> with </PROTEIN> BES1 </PROTEIN> or mutant bes1.
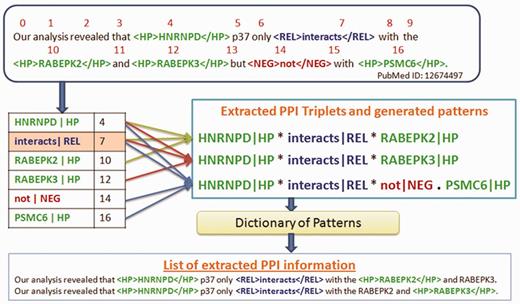
Algorithm to extract PPI triplets from complex sentences with more than two proteins.
| Rules . | Description . | Abstract Form 1 (PIP) . | Abstract Form 2 (IPP) . | Abstract Form 3 (PPI) . |
|---|---|---|---|---|
| Rule 1 | Order of two proteins and relation keyword | A | A | A |
| Rule 2 | Distance between the protein pair | NA | A | A |
| Rule 3 | Simple sentence with two proteins | A | A | A |
| Rule 4 | Simple sentence with two proteins and negation keyword | A | A | NA |
| Rule 5 | Complex sentence having more than two proteins | A | A | A |
| Rule 6 | Complex sentence having more than two proteins and negation keyword | A | A | NA |
| Rule 7 | Complex sentence having more than two proteins and two negation keyword | Special rule independent of Forms | ||
| Rules . | Description . | Abstract Form 1 (PIP) . | Abstract Form 2 (IPP) . | Abstract Form 3 (PPI) . |
|---|---|---|---|---|
| Rule 1 | Order of two proteins and relation keyword | A | A | A |
| Rule 2 | Distance between the protein pair | NA | A | A |
| Rule 3 | Simple sentence with two proteins | A | A | A |
| Rule 4 | Simple sentence with two proteins and negation keyword | A | A | NA |
| Rule 5 | Complex sentence having more than two proteins | A | A | A |
| Rule 6 | Complex sentence having more than two proteins and negation keyword | A | A | NA |
| Rule 7 | Complex sentence having more than two proteins and two negation keyword | Special rule independent of Forms | ||
PIP, protein–relation–protein; IPP, relation–protein–protein; PPI, protein–protein–relation; A, applicable; NA, not applicable
| Rules . | Description . | Abstract Form 1 (PIP) . | Abstract Form 2 (IPP) . | Abstract Form 3 (PPI) . |
|---|---|---|---|---|
| Rule 1 | Order of two proteins and relation keyword | A | A | A |
| Rule 2 | Distance between the protein pair | NA | A | A |
| Rule 3 | Simple sentence with two proteins | A | A | A |
| Rule 4 | Simple sentence with two proteins and negation keyword | A | A | NA |
| Rule 5 | Complex sentence having more than two proteins | A | A | A |
| Rule 6 | Complex sentence having more than two proteins and negation keyword | A | A | NA |
| Rule 7 | Complex sentence having more than two proteins and two negation keyword | Special rule independent of Forms | ||
| Rules . | Description . | Abstract Form 1 (PIP) . | Abstract Form 2 (IPP) . | Abstract Form 3 (PPI) . |
|---|---|---|---|---|
| Rule 1 | Order of two proteins and relation keyword | A | A | A |
| Rule 2 | Distance between the protein pair | NA | A | A |
| Rule 3 | Simple sentence with two proteins | A | A | A |
| Rule 4 | Simple sentence with two proteins and negation keyword | A | A | NA |
| Rule 5 | Complex sentence having more than two proteins | A | A | A |
| Rule 6 | Complex sentence having more than two proteins and negation keyword | A | A | NA |
| Rule 7 | Complex sentence having more than two proteins and two negation keyword | Special rule independent of Forms | ||
PIP, protein–relation–protein; IPP, relation–protein–protein; PPI, protein–protein–relation; A, applicable; NA, not applicable
PPI information extraction
Following the recognition of candidate PPI pairs based on three abstract forms and seven rules discussed above, the extraction of true PPIs from literature is a complicated and most challenging task because of the vast variations in grammatical structure of biomedical literature. To extract the true PPI and improve the accuracy, we constructed 11 specific patterns (four for abstract Form 1, three for abstract Form 2 and four for abstract Form 3) by mapping the semantic relations between the proteins combined with/without negation keywords for the three abstract forms. The 11 patterns are illustrated below using Tregex syntax (22) used in the Stanford parser package. The tags expressed in the syntax are listed in Table 2.
| Syntax tag . | Tag description . |
|---|---|
| S | Sentence |
| NP | Noun phrase |
| VP | Verb phrase |
| NNPS | Proper noun, plural |
| CC | Coordinating conjunction |
| IN | Preposition, subordinating conjunction |
| JJ | Adjective |
| DT | Determiner |
| $++ | Sister node on left |
| $+ | Immediate sisters |
| << | Points to root node |
| < | Points to next immediate node |
| PROTEIN1, PROTEIN2, PROTEIN3 | Special tag for protein |
| RELATION | Special tag for relation keyword |
| NEGATION | Special tag for negation keyword |
| And | Exact word match |
| With | Exact word match |
| Syntax tag . | Tag description . |
|---|---|
| S | Sentence |
| NP | Noun phrase |
| VP | Verb phrase |
| NNPS | Proper noun, plural |
| CC | Coordinating conjunction |
| IN | Preposition, subordinating conjunction |
| JJ | Adjective |
| DT | Determiner |
| $++ | Sister node on left |
| $+ | Immediate sisters |
| << | Points to root node |
| < | Points to next immediate node |
| PROTEIN1, PROTEIN2, PROTEIN3 | Special tag for protein |
| RELATION | Special tag for relation keyword |
| NEGATION | Special tag for negation keyword |
| And | Exact word match |
| With | Exact word match |
| Syntax tag . | Tag description . |
|---|---|
| S | Sentence |
| NP | Noun phrase |
| VP | Verb phrase |
| NNPS | Proper noun, plural |
| CC | Coordinating conjunction |
| IN | Preposition, subordinating conjunction |
| JJ | Adjective |
| DT | Determiner |
| $++ | Sister node on left |
| $+ | Immediate sisters |
| << | Points to root node |
| < | Points to next immediate node |
| PROTEIN1, PROTEIN2, PROTEIN3 | Special tag for protein |
| RELATION | Special tag for relation keyword |
| NEGATION | Special tag for negation keyword |
| And | Exact word match |
| With | Exact word match |
| Syntax tag . | Tag description . |
|---|---|
| S | Sentence |
| NP | Noun phrase |
| VP | Verb phrase |
| NNPS | Proper noun, plural |
| CC | Coordinating conjunction |
| IN | Preposition, subordinating conjunction |
| JJ | Adjective |
| DT | Determiner |
| $++ | Sister node on left |
| $+ | Immediate sisters |
| << | Points to root node |
| < | Points to next immediate node |
| PROTEIN1, PROTEIN2, PROTEIN3 | Special tag for protein |
| RELATION | Special tag for relation keyword |
| NEGATION | Special tag for negation keyword |
| And | Exact word match |
| With | Exact word match |
PPI patterns for abstract Form 1:
S ((NP << PROTEIN1) $++ (VP << RELATION) $++ (NP << PROTEIN2))
Example: PROTEIN1 interacts with PROTEIN2
S ((NP << PROTEIN1) $++ (VP << ((NP << RELATION) $++ (NP << PROTEIN2))))
Example: PROTEIN1 has weak association with PROTEIN2
S ((NP << PROTEIN1) $++ (VP << NEGATION $+ RELATION) $++ (NP << PROTEIN2))
Example: PROTEIN1 does not interact with PROTEIN2
S ((NP << PROTEIN1) $++ (VP << ((NP << NEGATION $+ RELATION) $++ (NP << PROTEIN2))))
Example: PROTEIN1 has no association with PROTEIN2
PPI patterns for abstract Form 2:
S (VP << RELATION $++ (NP << (PROTEIN1 $+ (CC < ‘and’) $+ PROTEIN2)))
Example: Interaction between PROTEIN1 and PROTEIN2
S (NP << RELATION $++ (NP << (PROTEIN1 $+ (IN < ‘with’) $+ PROTEIN2)))
Example: Interaction of PROTEIN1 with PROTEIN2
S (VP << (NEGATION $+ RELATION) $++ (NP << (PROTEIN1 $+ (CC < ‘and’) $+ PROTEIN2)))
Example: No detectable interaction between PROTEIN1 and PROTEIN2
Three independent patterns are defined for abstract Form 3, which itself is a pattern (h). A closer look at the biomedical literature expresses various forms of interacting protein pairs related to abstract Form 3: PROTEIN1/PROTEIN2, PROTEIN1-PROTEIN2 both correspond to pattern (i); PROTEIN1 and PROTEIN2 corresponds to pattern (j); PROTEIN1:PROTEIN2 corresponds to pattern (k). Presence of a negation keyword is not supported by this abstract form.
PPI patterns for abstract Form 3:
S (NP << PROTEIN1 $+ PROTEIN2 $+ (JJ < RELATION))
Example: PROTEIN1 PROTEIN2 complex
S (NP < (JJ < PROTEINS*) $+ (NN < RELATION))
Example: PROTEIN1/PROTEIN2 complex
S ((NP << PROTEIN1 $+ (CC < and) $+ PROTEIN2) $+ RELATION)
Example: PROTEIN1 and PROTEIN2 complex
S (NP << PROTEIN1 $++ PROTEIN2 $+ RELATION)
Example: PROTEIN1:PROTEIN2 complex
All the above 11 patterns are stored into a dictionary of patterns and applied for PPI information extraction. Figure 4 summarizes the extraction methodology of PPInterFinder.

Results and discussion
Datasets
Five standard corpora are available to evaluate PPI systems: AIMED (26), BioInfer (27), HPRD50 (28), IEPA (29) and LLL (30). All five corpora contain annotations for entities such as proteins and genes. Among these, AIMED and HPRD50 are specific to interactions related to human proteins. AIMED corpus comprises 200 PubMed abstracts containing PPI information and 25 abstracts without any PPI information as negative examples (26). HPRD50 is a sentence-based corpus containing 145 sentences with annotations and list of true and false PPI (28). We used AIMED and HPRD50 corpora to evaluate the performance of PPInterFinder as our system is specific to extract human PPIs.
In addition, we used our own dataset named as IntAct corpus, which was used to evaluate the performance of our system during BioCreative workshop 2012 (31). IntAct corpus consists of 693 sentences related to human proteins/genes interaction retrieved from the resource site of IntAct Database (ftp://ftp.ebi.ac.uk/pub/databases/intact/current/various/data-mining/). Furthermore, we use the PPInterFinder evaluation given by curators with their own datasets before and during BioCreative workshop 2012 at Washington DC, on 4–5 April 2012 (http://www.biocreative.org/tasks/bc-workshop-2012/Interactive_TM/).
Evaluation methods and metrics
Unlike other PPI systems, PPInterFinder is an integrated text mining tool with two in-built modules, a named entity tagging module known as NAGGNER (16) and protein/gene normalization module known as ProNormz (17). So, PPInterFinder can process and extract PPIs from raw text as well as text with pre-tagged protein/gene names.
Four different evaluations were conducted with PPInterFinder.
AIMED corpus specific to interactions related to human proteins
HPRD50 corpus specific to human proteins interactions
derived dataset from IntAct database with 693 sentences related to human proteins/genes interactions
Curators’ own dataset and evaluations provided by curators.
For (i), (ii) and (iii), the evaluations were carried out on raw text as well as text with tagged protein/gene names to compare the performance of PPInterFinder as an integrated text mining system (entity tagging, normalization and PPI extraction) and PPI extraction algorithm alone. For (iv), we used the evaluation results provided by the external curators of BioCreative workshop 2012.
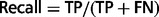
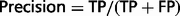
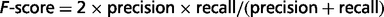
where TP (true positive) refers to the number or proportion of relations that were correctly extracted from input sentences; FN (false negative) refers to the number or proportion of relations that the system failed to extract from input sentences and FP (false positive) refers to the number of relations that were incorrectly extracted from input sentences. The F-score is the harmonic mean of recall and precision.
Evaluation on AIMED, HPRD50, IntAct corpora
The AIMED corpus consists of 200 PubMed abstracts from DIP (6) with known PPI information (26). These abstracts were manually annotated for interactions between human genes/proteins. In addition, 25 abstracts without any PPI information are added to the corpus as negative examples (Supplementary Data 2). The HPRD50 corpus was created from 50 abstracts referenced by the Human Protein Reference Database (HPRD) (28). The annotated genes/proteins entities of the corpus include 266 relation instances (i.e. pairs of genes/proteins), corresponding to 126 direct physical relations and 35 regulatory relations (Supplementary Data 3). The IntAct corpus consists of 693 sentences related to human proteins/genes interactions and was manually curated by us (Supplementary Data 4).
We performed two types of evaluation, i.e. text with pre-tagged protein/gene names as well as raw text using these three corpora as mentioned earlier. Table 3 shows the results of PPInterFinder on the three corpora.
| Corpus . | AIMED . | HPRD50 . | IntAct . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . |
| PPI algorithm | ||||||||||||||||||
| PIP | 432 | 72 | 233 | 64.96 | 85.71 | 73.91 | 73 | 7 | 49 | 59.84 | 91.25 | 72.28 | 270 | 34 | 73 | 78.72 | 88.82 | 83.47 |
| IPP | 103 | 39 | 137 | 42.92 | 69.13 | 52.96 | 9 | 5 | 15 | 37.50 | 64.29 | 47.37 | 64 | 12 | 33 | 65.98 | 84.21 | 73.99 |
| PPI | 42 | 31 | 81 | 34.15 | 57.53 | 42.85 | 5 | 1 | 4 | 55.56 | 83.33 | 66.67 | 70 | 20 | 24 | 74.47 | 77.78 | 76.09 |
| Total | 577 | 142 | 451 | 56.12 | 80.25 | 66.05 | 87 | 13 | 68 | 56.13 | 87.00 | 68.24 | 404 | 55 | 130 | 75.66 | 88.01 | 81.37 |
| PPI algorithm with preprocessing steps (NER and GN) | ||||||||||||||||||
| PIP | 334 | 68 | 331 | 50.23 | 83.08 | 62.61 | 49 | 5 | 75 | 39.52 | 90.74 | 55.04 | 258 | 34 | 85 | 75.22 | 88.36 | 81.26 |
| IPP | 95 | 41 | 144 | 39.75 | 69.85 | 50.65 | 8 | 6 | 17 | 32.00 | 57.14 | 41.02 | 53 | 12 | 44 | 54.64 | 81.54 | 65.43 |
| PPI | 40 | 16 | 96 | 29.41 | 71.43 | 41.67 | 3 | 1 | 6 | 50.00 | 75.00 | 60.00 | 65 | 20 | 29 | 69.15 | 76.47 | 72.63 |
| Total | 469 | 125 | 571 | 45.10 | 78.96 | 57.41 | 60 | 12 | 98 | 37.97 | 83.33 | 52.17 | 376 | 55 | 158 | 70.58 | 87.33 | 78.07 |
| Corpus . | AIMED . | HPRD50 . | IntAct . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . |
| PPI algorithm | ||||||||||||||||||
| PIP | 432 | 72 | 233 | 64.96 | 85.71 | 73.91 | 73 | 7 | 49 | 59.84 | 91.25 | 72.28 | 270 | 34 | 73 | 78.72 | 88.82 | 83.47 |
| IPP | 103 | 39 | 137 | 42.92 | 69.13 | 52.96 | 9 | 5 | 15 | 37.50 | 64.29 | 47.37 | 64 | 12 | 33 | 65.98 | 84.21 | 73.99 |
| PPI | 42 | 31 | 81 | 34.15 | 57.53 | 42.85 | 5 | 1 | 4 | 55.56 | 83.33 | 66.67 | 70 | 20 | 24 | 74.47 | 77.78 | 76.09 |
| Total | 577 | 142 | 451 | 56.12 | 80.25 | 66.05 | 87 | 13 | 68 | 56.13 | 87.00 | 68.24 | 404 | 55 | 130 | 75.66 | 88.01 | 81.37 |
| PPI algorithm with preprocessing steps (NER and GN) | ||||||||||||||||||
| PIP | 334 | 68 | 331 | 50.23 | 83.08 | 62.61 | 49 | 5 | 75 | 39.52 | 90.74 | 55.04 | 258 | 34 | 85 | 75.22 | 88.36 | 81.26 |
| IPP | 95 | 41 | 144 | 39.75 | 69.85 | 50.65 | 8 | 6 | 17 | 32.00 | 57.14 | 41.02 | 53 | 12 | 44 | 54.64 | 81.54 | 65.43 |
| PPI | 40 | 16 | 96 | 29.41 | 71.43 | 41.67 | 3 | 1 | 6 | 50.00 | 75.00 | 60.00 | 65 | 20 | 29 | 69.15 | 76.47 | 72.63 |
| Total | 469 | 125 | 571 | 45.10 | 78.96 | 57.41 | 60 | 12 | 98 | 37.97 | 83.33 | 52.17 | 376 | 55 | 158 | 70.58 | 87.33 | 78.07 |
Performance evaluation (%): recall (R), precision (P) and F-score (F); NER, named entity recognition; GN, gene/protein normalization.
| Corpus . | AIMED . | HPRD50 . | IntAct . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . |
| PPI algorithm | ||||||||||||||||||
| PIP | 432 | 72 | 233 | 64.96 | 85.71 | 73.91 | 73 | 7 | 49 | 59.84 | 91.25 | 72.28 | 270 | 34 | 73 | 78.72 | 88.82 | 83.47 |
| IPP | 103 | 39 | 137 | 42.92 | 69.13 | 52.96 | 9 | 5 | 15 | 37.50 | 64.29 | 47.37 | 64 | 12 | 33 | 65.98 | 84.21 | 73.99 |
| PPI | 42 | 31 | 81 | 34.15 | 57.53 | 42.85 | 5 | 1 | 4 | 55.56 | 83.33 | 66.67 | 70 | 20 | 24 | 74.47 | 77.78 | 76.09 |
| Total | 577 | 142 | 451 | 56.12 | 80.25 | 66.05 | 87 | 13 | 68 | 56.13 | 87.00 | 68.24 | 404 | 55 | 130 | 75.66 | 88.01 | 81.37 |
| PPI algorithm with preprocessing steps (NER and GN) | ||||||||||||||||||
| PIP | 334 | 68 | 331 | 50.23 | 83.08 | 62.61 | 49 | 5 | 75 | 39.52 | 90.74 | 55.04 | 258 | 34 | 85 | 75.22 | 88.36 | 81.26 |
| IPP | 95 | 41 | 144 | 39.75 | 69.85 | 50.65 | 8 | 6 | 17 | 32.00 | 57.14 | 41.02 | 53 | 12 | 44 | 54.64 | 81.54 | 65.43 |
| PPI | 40 | 16 | 96 | 29.41 | 71.43 | 41.67 | 3 | 1 | 6 | 50.00 | 75.00 | 60.00 | 65 | 20 | 29 | 69.15 | 76.47 | 72.63 |
| Total | 469 | 125 | 571 | 45.10 | 78.96 | 57.41 | 60 | 12 | 98 | 37.97 | 83.33 | 52.17 | 376 | 55 | 158 | 70.58 | 87.33 | 78.07 |
| Corpus . | AIMED . | HPRD50 . | IntAct . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . | TP . | FP . | FN . | R . | P . | F . |
| PPI algorithm | ||||||||||||||||||
| PIP | 432 | 72 | 233 | 64.96 | 85.71 | 73.91 | 73 | 7 | 49 | 59.84 | 91.25 | 72.28 | 270 | 34 | 73 | 78.72 | 88.82 | 83.47 |
| IPP | 103 | 39 | 137 | 42.92 | 69.13 | 52.96 | 9 | 5 | 15 | 37.50 | 64.29 | 47.37 | 64 | 12 | 33 | 65.98 | 84.21 | 73.99 |
| PPI | 42 | 31 | 81 | 34.15 | 57.53 | 42.85 | 5 | 1 | 4 | 55.56 | 83.33 | 66.67 | 70 | 20 | 24 | 74.47 | 77.78 | 76.09 |
| Total | 577 | 142 | 451 | 56.12 | 80.25 | 66.05 | 87 | 13 | 68 | 56.13 | 87.00 | 68.24 | 404 | 55 | 130 | 75.66 | 88.01 | 81.37 |
| PPI algorithm with preprocessing steps (NER and GN) | ||||||||||||||||||
| PIP | 334 | 68 | 331 | 50.23 | 83.08 | 62.61 | 49 | 5 | 75 | 39.52 | 90.74 | 55.04 | 258 | 34 | 85 | 75.22 | 88.36 | 81.26 |
| IPP | 95 | 41 | 144 | 39.75 | 69.85 | 50.65 | 8 | 6 | 17 | 32.00 | 57.14 | 41.02 | 53 | 12 | 44 | 54.64 | 81.54 | 65.43 |
| PPI | 40 | 16 | 96 | 29.41 | 71.43 | 41.67 | 3 | 1 | 6 | 50.00 | 75.00 | 60.00 | 65 | 20 | 29 | 69.15 | 76.47 | 72.63 |
| Total | 469 | 125 | 571 | 45.10 | 78.96 | 57.41 | 60 | 12 | 98 | 37.97 | 83.33 | 52.17 | 376 | 55 | 158 | 70.58 | 87.33 | 78.07 |
Performance evaluation (%): recall (R), precision (P) and F-score (F); NER, named entity recognition; GN, gene/protein normalization.
The reported F-scores of AIMED, HPRD50 and IntAct corpora on tagged text were 66.05, 68.24 and 81.37 and on raw text were 57.41, 52.17 and 78.07, respectively. The lower F-score achieved by our system on two standard corpora AIMED and HPRD50 was because of lower recall (Table 3). This is due to the presence of more than one relation keyword (135 sentences in AIMED and 32 sentences in HPRD50) per sentence and PPI information spread across the sentences as in AIMED corpus. However, human curated IntAct corpus contains sentences with one relation keyword and no PPI information spread across the sentence boundaries. Subsequently, our system achieves higher recall of 75.66 on tagged text and 70.58 on raw text and achieves a higher F-score on both evaluations. PPI extractions from sentences having more than one relation keyword and relations across the sentences are future objectives of PPInterFinder.
Table 3 also presents the evaluation results for the three abstract forms in three corpora both on tagged text and raw text. The important benefit of such an evaluation is to understand the performance of PPInterFinder on each abstract forms and their distribution in all three corpora. Abstract Form 1 achieves higher F-score values of 73.91, 72.28 and 83.47 on AIMED, HPRD50 and IntAct corpora, respectively, on tagged text. This result clearly demonstrates that the performance of our PPI extraction algorithm is comparatively better on abstract Form 1. In our three test corpora, we observed that the number of tokens between the protein pair and the relation keyword vary widely in abstract Forms 2 and 3. We fixed the number of tokens to one for abstract Form 2 and one or no token for abstract Form 3 (Rule 2) to reduce the extraction of many false PPI pairs. Consequently, few PPI pairs with more than one token between the protein pairs and the relation keyword remain unidentified and report for lower F-score on abstract Forms 2 and 3. In addition, Table 3 clearly shows that abstract Form 1 is the most common one in all three corpora, accounting for maximum number of TP + FN value, i.e. 665 on AIMED, 122 in HPRD50 and 343 in IntAct corpora. The other two abstract forms were comparatively less common in all three corpora.
Table 3 also shows the results of PPI extraction algorithm with/without the preprocessing steps on the three corpora. The reported accuracy of our protein/gene tagging system NAGGNER was 75.77% (16), which was equivalent to other state of the art biomedical NER systems (32). It was obvious from our results that if we use the raw text, the performance of PPInterFinder was decreased to the overall F-score of 5–10% in all the three corpora as few genes/proteins remain unidentified and not tagged in the preprocessing steps. We are the first one to report the decrease in performance of 5–10% if the raw text is used for PPI task and it would be the problem of interest to investigate further.
Negation keyword recognition is another additional feature of PPInterFinder. Presence of any negation keyword in a sentence confirms that two genes/proteins do not interact. PPInterFinder recognizes the presence of ‘no’, ‘not’ and ‘neither/nor’ as negation keywords for false PPI information. Surprisingly, evaluation on the three corpora AIMED, HPRD50 and IntAct confirms that they contain very few sentences with negation keyword (five in AIMED, two in HPRD50 and three in IntAct). These results indicate that the negation keyword recognition will not affect the overall performance of the PPI system but it is helpful to exclude few false PPIs.
Direct comparison of our system with others is not possible, as PPInterFinder is exclusively developed to extract human PPIs. However, we utilized the comparison table of different PPI systems given by Bui et al. (18) on AIMED corpus as it is specific to human proteins. Comparison of our system with the existing systems on AIMED corpus is given in Table 4. PPInterFinder achieves a highest F-score of 66.05 against others. The highest F-score by PPInterFinder is due the following facts:
Rich set of relation keywords specific to human proteins (Supplementary Data 1)
Parser with seven rules to identify candidate PPI pairs
True PPI information extraction using 11 patterns specific to the syntactic structure of the biomedical sentence
| System . | Description . | F-score (%) . |
|---|---|---|
| Saetre et al. (33) | Feature-based, two parsers | 64.2 |
| Miwa et al. (34) | Multiple kernels, two parsers | 60.8 |
| Kim et al. (35) | Walk-weighted subsequence kernels, one parser | 56.6 |
| Airola et al. (36) | All-paths graph kernel, one parser | 56.4 |
| Niu et al. (14) | All-paths graph kernel, one parser | 53.5 |
| Bui et al. (18) | RBF kernel, one parser | 61.2 |
| PPInterFinder | Pattern matching, two parsers | 66.05 |
| System . | Description . | F-score (%) . |
|---|---|---|
| Saetre et al. (33) | Feature-based, two parsers | 64.2 |
| Miwa et al. (34) | Multiple kernels, two parsers | 60.8 |
| Kim et al. (35) | Walk-weighted subsequence kernels, one parser | 56.6 |
| Airola et al. (36) | All-paths graph kernel, one parser | 56.4 |
| Niu et al. (14) | All-paths graph kernel, one parser | 53.5 |
| Bui et al. (18) | RBF kernel, one parser | 61.2 |
| PPInterFinder | Pattern matching, two parsers | 66.05 |
| System . | Description . | F-score (%) . |
|---|---|---|
| Saetre et al. (33) | Feature-based, two parsers | 64.2 |
| Miwa et al. (34) | Multiple kernels, two parsers | 60.8 |
| Kim et al. (35) | Walk-weighted subsequence kernels, one parser | 56.6 |
| Airola et al. (36) | All-paths graph kernel, one parser | 56.4 |
| Niu et al. (14) | All-paths graph kernel, one parser | 53.5 |
| Bui et al. (18) | RBF kernel, one parser | 61.2 |
| PPInterFinder | Pattern matching, two parsers | 66.05 |
| System . | Description . | F-score (%) . |
|---|---|---|
| Saetre et al. (33) | Feature-based, two parsers | 64.2 |
| Miwa et al. (34) | Multiple kernels, two parsers | 60.8 |
| Kim et al. (35) | Walk-weighted subsequence kernels, one parser | 56.6 |
| Airola et al. (36) | All-paths graph kernel, one parser | 56.4 |
| Niu et al. (14) | All-paths graph kernel, one parser | 53.5 |
| Bui et al. (18) | RBF kernel, one parser | 61.2 |
| PPInterFinder | Pattern matching, two parsers | 66.05 |
Evaluation by Biocurators before and during BioCreative Workshop 2012
Prior to the Workshop, two curators from PPI databases BioGrid and MINT evaluated the system with a set of 50 abstracts related to human proteins with the main focus on human protein kinases (Supplementary Data 5). The performance of PPInterFinder was evaluated in two stages similar to our evaluation on other three corpora, namely (i) based on PPI extraction algorithm alone and (ii) based on PPI extraction algorithm including preprocessing steps. The reported F-scores were 76.91 for the tagged text and 60.61 for raw text by curator 1 and 73.17 for tagged text and 60.61 for raw text by curator 2 (Table 5). The difference in F-score between the two curators was mainly due to their manual annotation (46 PPIs identified by curator1 and 52 PPIs by curator2) (Supplementary Data 5).
| Evaluation . | Curator1 . | Curator2 . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| Preprocessing steps (NER & GN) + PPI extraction algorithm | 46.88 | 85.71 | 60.61 | 46.88 | 85.71 | 60.61 |
| PPI extraction algorithm | 69.76 | 85.71 | 76.91 | 63.83 | 85.71 | 73.17 |
| Evaluation . | Curator1 . | Curator2 . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| Preprocessing steps (NER & GN) + PPI extraction algorithm | 46.88 | 85.71 | 60.61 | 46.88 | 85.71 | 60.61 |
| PPI extraction algorithm | 69.76 | 85.71 | 76.91 | 63.83 | 85.71 | 73.17 |
Performance evaluation (%): recall (R), precision (P) and F-score (F); NER, named entity recognition; GN, gene/protein normalization.
| Evaluation . | Curator1 . | Curator2 . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| Preprocessing steps (NER & GN) + PPI extraction algorithm | 46.88 | 85.71 | 60.61 | 46.88 | 85.71 | 60.61 |
| PPI extraction algorithm | 69.76 | 85.71 | 76.91 | 63.83 | 85.71 | 73.17 |
| Evaluation . | Curator1 . | Curator2 . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| Preprocessing steps (NER & GN) + PPI extraction algorithm | 46.88 | 85.71 | 60.61 | 46.88 | 85.71 | 60.61 |
| PPI extraction algorithm | 69.76 | 85.71 | 76.91 | 63.83 | 85.71 | 73.17 |
Performance evaluation (%): recall (R), precision (P) and F-score (F); NER, named entity recognition; GN, gene/protein normalization.
In addition, the performance of PPInterFinder was evaluated by three additional curators during the workshop at Washington DC, on 4–5 April 2012 (http://www.biocreative.org/tasks/bc-workshop-2012/Interactive_TM/). This was an informal evaluation comprising only the subjective measure on a set of survey questionnaires. The system was rated under six main categories, namely, overall reaction, system’s ability to help complete tasks, design of application, learning to use the application, usability and finally recommendation of the system. While two curators (1 and 3) have recommended the system as 4 (maximum score is 7), curator 2 suggested to decrease the number of false positives from the reported value of 88 (Supplementary Data 6).
Improvements after BioCreative workshop 2012
During BioCreative workshop 2012, the system was evaluated only with the derived dataset of 693 sentences from IntAct database and the reported accuracy was 75.94% (31). The curators reported the extraction of 88 false PPIs (false positive) by PPInterFinder were mainly due to the inclusion of some common relation keywords (e.g. add, contain, increase, reduce and localize). In the present improved version, we modified the PPI extraction methodology by incorporating the following three major updates:
Twenty-one relation keywords related to the above five relation keywords groups were removed from the relation keyword dictionary as these keywords extract many false PPIs than true PPIs. For example, the relation keyword ‘addition’ extracting false PPI information is illustrated below.
Example 4:
PubMed ID: 18001825: In <RELATION> addition </RELATION>, <PROTEIN> RNF8 </PROTEIN> coprecipitated with Del mutant of <PROTEIN> MDC1 </PROTEIN> in vivo.
We introduced two new rules (Rules 1 and 2) for checking the position of relation keyword with a pair of proteins and the number of tokens between the proteins in the candidate PPI pair identification phase.
We added the true PPI extraction methodology by incorporating 11 specific patterns related to the three abstract forms.
We tested the performance of the updated algorithm with IntAct corpus (Supplementary Data 6). The number of false positives was reduced to 55 in the improved version, with the overall F-score of 78.07%. The improved performance is shown in Table 6. Manual analysis on the list of 55 false positives confirms that one or more proteins remain unidentified in 30 sentences in the preprocessing steps. Consequently the extracted information is a false PPI (Supplementary Data 7). Figure 5 shows the input and the extracted output of PPInterFinder.
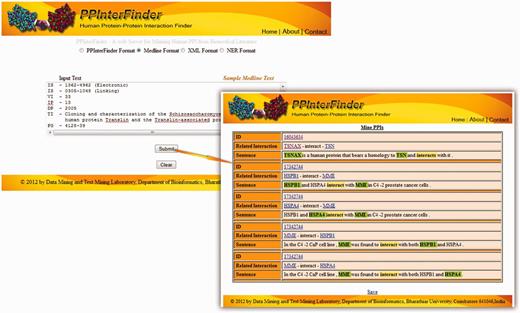
Screenshot of PPInterFinder showing input and extracted PPI pairs.
| Dataset . | PPInterFinder (improved version) . | PPInterFinder (BioCreative Workshop 2012) . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| 693 sentences from IntAct Database | 70.58 | 87.33 | 78.07 | 71.27 | 81.28 | 75.94 |
| Dataset . | PPInterFinder (improved version) . | PPInterFinder (BioCreative Workshop 2012) . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| 693 sentences from IntAct Database | 70.58 | 87.33 | 78.07 | 71.27 | 81.28 | 75.94 |
Performance evaluation (%): recall (R), precision (P) and F-score (F).
| Dataset . | PPInterFinder (improved version) . | PPInterFinder (BioCreative Workshop 2012) . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| 693 sentences from IntAct Database | 70.58 | 87.33 | 78.07 | 71.27 | 81.28 | 75.94 |
| Dataset . | PPInterFinder (improved version) . | PPInterFinder (BioCreative Workshop 2012) . | ||||
|---|---|---|---|---|---|---|
| . | R . | P . | F . | R . | P . | F . |
| 693 sentences from IntAct Database | 70.58 | 87.33 | 78.07 | 71.27 | 81.28 | 75.94 |
Performance evaluation (%): recall (R), precision (P) and F-score (F).
Conclusion
We have developed an integrated text mining system PPInterFinder for extracting causal relations between human proteins by applying a set of rules on grammatically parsed sentence to identify the candidate PPI pairs and matching the syntactic structure of the sentence with a dictionary of patterns. To our knowledge, PPInterFinder is the only system that integrates two preprocessing modules, protein/gene name tagging and normalization. Hence, PPInterFinder handles raw text as well as pre-tagged text as per user requirement. The evaluation of PPInterFinder on four benchmarked corpora has shown that our system achieves results comparable with other best PPI extraction methods and further, there is a decrease in overall F-score of 5–10% when gold standard NER text is not used. We are the first one to report this. In present form, the system is available for human PPI information extraction on single sentences with two or more proteins and one relation keyword. The extraction of PPI information across the sentences and on sentences having multiple relation keywords are the future objectives of PPInterFinder.
Funding
Department of Information Technology (DIT), Government of India. [DIT/R&D/BIO/15(22)/2008]. KR and SS acknowledge the fellowships received from the grant. Funding for open access charge: DIT and Bharathiar University.
Conflict of interest. None declared.
References
Author notes
Citation details: Kalpana Raja, Suresh Subramani, and Jeyakumar Natarajan. PPInterFinder—a mining tool for extracting causal relations on human proteins from literature. Database (2013) Vol. 2013: article ID bas052; doi:10.1093/database/bas052.



