-
PDF
- Split View
-
Views
-
Cite
Cite
Chengyu Wang, Tingting Chen, Yuchen Mu, Xuan Liang, Kai Xiong, Liqiang Ai, Yunyan Gu, Xingxing Fan, Haihai Liang, FDRdb: a manually curated database of fibrotic disease–associated RNAome and high-throughput datasets, Database, Volume 2022, 2022, baac095, https://doi.org/10.1093/database/baac095
Close - Share Icon Share
Abstract
Fibrosis is a common and serious disease that exists as a complicated impairment in many organs and triggers a complex cascade of responses. The deregulation of Ribonucleic Acids (RNAs) plays important roles in a variety of organ fibrosis cases. However, for fibrotic diseases, there is still a lack of an integrated platform with up-to-date information on RNA deregulation and high-throughput data. The Fibrotic Disease–associated RNAome database (FDRdb) (http://www.medsysbio.org/FDRdb) is a manually curated database of fibrotic disease–associated RNAome information and high-throughput datasets. This initial release (i) contains 1947 associations between 912 RNAs and 92 fibrotic diseases in eight species; (ii) collects information on 764 datasets of fibrotic diseases; (iii) provides a user-friendly web interface that allows users to browse, search and download the RNAome information on fibrotic diseases and high-throughput datasets and (iv) provides tools to analyze the expression profiles of fibrotic diseases, including differential expression analysis and pathway enrichment. The FDRdb is a valuable resource for researchers to explore the mechanisms of RNA dysregulation in organ fibrosis.
Database URL: http://www.medsysbio.org/FDRdb
Introduction
Fibrosis is a common pathological outcome of severe tissue injury or the dysregulation of wound-healing repair. Fibrotic disease is defined as the abnormal deposition of the extracellular matrix, and it involves various organs, such as the lung, liver, kidney, heart and skin (1–3). For example, idiopathic pulmonary fibrosis (IPF) is a chronic progressive disease characterized by destroyed pulmonary alveolar architecture (4, 5). Other fibrotic diseases similar to IPF, such as cirrhosis and heart failure, also cause organ damage, yet their mechanisms are still unclear (6, 7). Because fibrotic diseases are remarkably complex and induced by multiple pathogenic factors, uncovering the mechanism behind fibrosis is the primary challenge in organ fibrosis research.
The RNAome involved in fibrotic diseases includes two categories: coding (RNA) and non-coding RNA (ncRNA). Coding RNAs encode proteins, whereas ncRNAs include long ncRNAs (lncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs) and small-interfering RNAs (siRNAs). Recently, increasing evidence has suggested that alterations in RNA molecules play critical roles in the progression of fibrotic diseases. For example, miR-21 is a mediator involved in the pathogenesis of cardiac fibrosis, and the pharmacological silencing of miR-21 may be a potential therapeutic approach for treating cardiomyopathy associated with Chagas disease (8). Furthermore, longitudinally stable serum chitinase 1 activity and YKL-40 concentration levels may potentially be associated with the antifibrotic treatment response in IPF (9). LncRNA growth arrest specific 5 (GAS5) was decreased in the cirrhotic liver, and the down-regulation of GAS5 could inhibit cirrhosis (10). High glucose–induced exo-circ_0125310 promoted cell proliferation and fibrosis in diabetic nephropathy via sponging miR-422a and targeting the insulin like growth factor 1 receptor/p38 axis (11). Moreover, CD98 siRNA-loaded nanoparticles can decrease hepatic steatosis in mice (12). Currently, many experiments have confirmed associations between the dysregulation of RNAs and fibrotic diseases, which are accompanied by a large number of relevant datasets. However, these data are presented by different publications, making it difficult for researchers to access them. Therefore, it is worthwhile to construct a database to integrate all the published information on RNAs and the datasets for organ fibrosis.
Several databases have been built to collect and organize disease–related information on RNA. For example, the Nervous System Disease NcRNAome Atlas focuses on associations between nervous system diseases and ncRNAs (13), whereas the Pancreatic Expression Database is one of the main repositories for pancreatic-derived-omics data (14). Furthermore, the CTdatabase provides basic genome information concerning testis cancer (15). However, a high-quality curated, fibrotic disease–related RNA public resource, including high-throughput datasets, remains unavailable.
To bridge this gap, we developed a database, called the Fibrotic Disease–associated RNAome database (FDRdb) (http://www.medsysbio.org/FDRdb), which serves as a manually curated, open, up-to-date and online-based depository of RNA information and datasets about fibrotic diseases across various species.
The current version of the FDRdb documents 1947 associations between 912 RNAs and 92 fibrotic diseases in eight species, which were manually collected from 957 pieces of the published literature. The FDRdb also has 764 fibrotic disease datasets for researchers to browse and download, which includes 624 RNA expression profiles, 32 methylation profiles and 108 sequencing datasets. Meanwhile, the FDRdb allows users to upload their own RNA expression profile to perform differential expression analyses and pathway enrichment. The FDRdb could allow researchers to further investigate the relationship between RNAs and fibrotic diseases.
Materials and methods
Data collection
To collect as many fibrotic disease–associated RNAs as possible, all FDRdb entries were manually collected through several public databases. The naming of fibrotic diseases was done in keeping with the principle of the Medical Subject Heading disease categories, which contains 82 fibrotic diseases (16). Protein-coding genes were collected from the National Center for Biotechnology Information (NCBI) database (17). The ncRNA information was integrated from various resources: lncRNAs were downloaded from lncRNAdb (18); NONCODE (19); mature and precursor miRNAs with IDs were downloaded from miRbase (20) and circRNAs were collected from circBase (21). All gene symbols used in the FDRdb conformed to the HUGO Gene Nomenclature Committee guidelines. Thereafter, we used a simple script to obtain all abstracts in the PubMed database using keyword combinations, such as ‘fibrotic disease’ and/or ‘RNA symbol’. Subsequently, we covered all published pieces of literature describing the associations between RNAs and fibrotic diseases. Finally, we manually extracted credible entries from tens of thousands of literature pieces. We also derived fibrotic disease–related RNAs from other databases, including LncRNADisease 2.0 (22), Circad (23), circRNADisease (24) and miR2Disease (25). All the data from other public databases should comply with the appropriate naming standards and have been compared with the data we manually collected. The redundant entries stored in the FDRdb were removed.
Because different RNA interaction patterns (such as messenger RNA (mRNA)–mRNA, mRNA–lncRNA and miRNA–lncRNA) are involved in fibrotic diseases, we hypothesized that the RNA–RNA interactions in which both RNAs were related to fibrotic diseases may contribute to these diseases. We curated RNA interaction data from public databases, such as the Molecular Signatures Database (MsigDB) v3.0 (26) and RNAInter v4.0 (27). Finally, 1063 RNA–RNA interactions were predicted with a high potentiality of involving fibrosis disease.
The datasets of fibrotic disease–associated RNAs are another part of the FDRdb. We searched the fibrotic disease datasets using related keywords (e.g. ‘liver fibrosis, miRNA’) in public databases, including Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home), BioProject (https://www.ncbi.nlm.nih.gov/bioproject) and ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and collected detailed information for each dataset. All selected entries were further inspected and corrected by at least two researchers.
Confidence score
To evaluate how closely RNA is associated with the fibrotic disease, we provided a confidence score. The confidence score is calculated based on four factors: (i) biological and experimental verification (S1 = {1, 0}); (ii) medication information (S2 = {1, 0}); (iii) information on both RNA and pathway, either RNA or pathway (S3 = {1, 0.5, 0}) and (iv) the number of the published studies speaking on this association (n). The calculation formula is as follows:
To help users to better screen target associations, we set the threshold as follows: the associations with a confidence score between 0 and 1 are weak correlations, those between 1 and 3 are medium correlations and those greater than 3 are strong correlations.
Database construction
Linux, Apache, MySQL and Python (LAMP) architecture were implemented to build the dynamic database backend. The FDRdb was built on the Apache HTTP server in the CentOS operating system. All FDRdb data were stored and managed using MySQL. The web was developed with Django and HTML, and the web interface was built using HTML/CSS/JS. The FDRdb has been tested in Google Chrome (version 99.0.4844.84) and Firefox (version 101.0.1) browsers. All analysis functions provided by the FDRdb were based on Python software version 3.9.13.
Results
Database content
After sifting through and compiling all the entries we found from public sources and the literature, the FDRdb had 1947 RNA entries about 92 fibrotic diseases in eight species, as well as 377 predicted RNA–RNA interactions that contributed to fibrotic diseases, and 357 fibrotic disease–associated datasets. For each entry of RNA-associated fibrotic diseases, the FDRdb contains detailed information about the name of the fibrotic disease (e.g. liver cirrhosis), the expression pattern (e.g. up-regulation, down-regulation), species (e.g. Homo sapiens), tissue/cell line (e.g. lung, liver), the detection methods (e.g. RNA-seq, Real-Time Quantitative Reverse Transcription PCR (qRT-PCR), microarray), molecular target, pathway and fibrotic disease–related function. Each entry also has the title and public year of the corresponding literature together with a PubMed ID and a hyper link to the NCBI PubMed database (28). For each entry of datasets, the FDRdb provides the database name, data ID, data type (e.g. expression), platform, disease name, tissue, species, sample number, keyword and basic data description. Additionally, we offer a download link to help researchers to conveniently access the original data’s download page.
User interface
The FDRdb provides a user-friendly web interface that allows users to browse, search, analyze and download (Figure 1). The ‘Browse’ page provides users with two options: browse by RNA or browse by dataset. In the ‘RNA browse’ page, the ‘optional tree’ contains five types of RNAs, eight types of species, 92 types of diseases and 15 types of tissues (Figure 1A). Furthermore, it provides a list of fibrotic diseases that have been reported to be associated with COVID-19. In the ‘Dataset browse’ page, the ‘optional tree’ contains four categories: three kinds of data types, eight types of species, 69 types of diseases, and nine types of tissues. Users can select the entries under the categories of interest to view specific information about fibrosis. The ‘Search’ page allows users to search corresponding fibrosis-associated information by typing a keyword (Figure 1B). Users can search using different types of keywords: mRNA symbol, miRNA symbol, lncRNA symbol, circRNA symbol, siRNA symbol or fibrotic disease. The ‘Search’ page also provides users with pull-down menus to select all or specific species or tissues. Some examples for searching are provided on the bottom of the interface on this page to help users to navigate. On the result page, the FDRdb shows the entries in a table (Figure 1C). Users can then select the ‘click here to download the table’ option. Furthermore, users can obtain more detailed information by clicking on the ‘Details’ link (Figure 1D). On the ‘Data Details’ page, users can jump to the original source to download the fibrotic dataset by clicking the ‘Download’ button (Figure 1E). The ‘Link’ page lists the external hyperlink of the public databases and data resources used in the FDRdb. The ‘Download’ page allows users to download all data concerning the relationship between the fibrotic diseases and RNAs in the FDRdb. The fibrotic diseases-miRNA, -lncRNA, -mRNA, -circRNA and -siRNA association data may be downloaded in .txt or .csv formats. The FDRdb also allows users to download data on the fibrotic disease–RNA relationship by choosing a specific species, such as H. sapiens, Mus musculus or Rattus norvegicus.
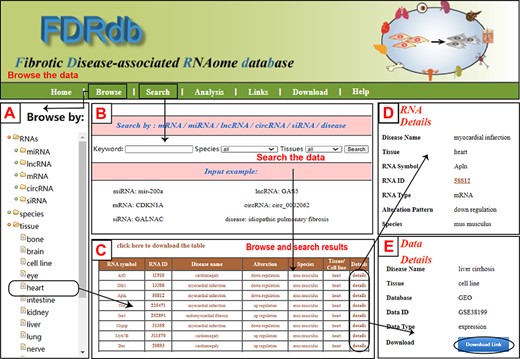
A schematic workflow of the FDRdb. (A) The ‘RNA browse’ page. (B) The ‘Search’ page. (C) The ‘Result’ page. (D) The detailed information of RNA entries. (E) The detailed information of dataset entries.
The FDRdb allows users to analyze their own RNA expression profile. On the ‘Analysis’ page, users can upload expression profile and sample classification information to perform differential RNA expression analysis. Moreover, users can select specific species to get corresponding differentially expressed fibrotic RNAs between two types of samples (Figure 2A). The graph shows the differentially expressed RNA information between two types of samples and highlights whether these differentially expressed RNAs are involved in some specific fibrotic disease. Using the example files provided by the FDRdb, when users click the ‘Apply’ button, the FDRdb will navigate to the results page where a brief summary of the results is listed on the top of the page (Figure 2B). Meanwhile, the FDRdb visualizes the analysis results, which include the volcano plot of differential RNAs (|log(fold-change)| > 1 and T-test, P < 0.05; Figure 2C), the hierarchical clustering heatmap of differentially expressed RNAs based on the Euclidean distance matrix (Figure 2D), the top 20 significantly enriched pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG) (Hypergeometric test, P < 0.05; Figure 2E) and biological processes in Gene Ontology (GO) (Hypergeometric test, P < 0.05; Figure 2F). All results and figures can be downloaded.
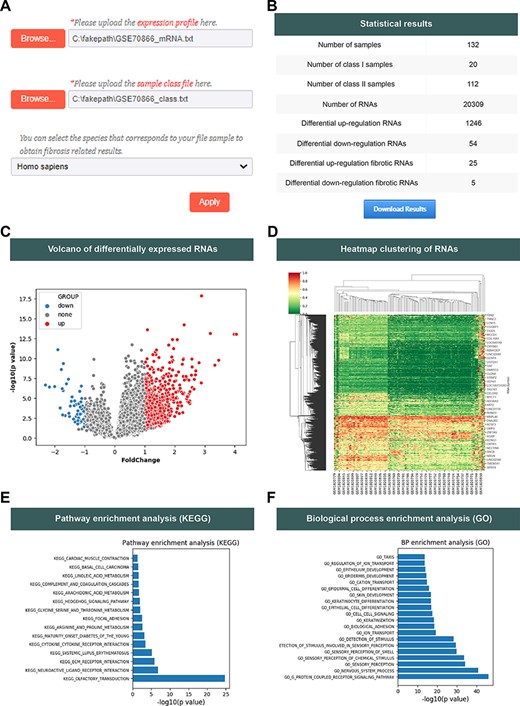
The analysis function of the FDRdb. (A) The upload interface on the ‘Analysis’ page. (B) The statistics of analysis results. (C) The volcano plot of differentially expressed RNAs. (D) The heatmap clustering of differentially expressed RNAs. A statistical bar chart of pathway enrichment analysis in KEGG (E) and analysis of biological process enrichment in GO (F).
We also provided a detailed tutorial on the ‘Help’ page, where users can also get statistics about the data stored in the FDRdb.
Landscape of the data in the FDRdb
To have a landscape of the associated data in the FDRdb, we constructed a network to show the distribution of associations between RNAs and fibrotic diseases in different tissues (Figure 3A). By analyzing the topology of the network, we found that the lung, heart, kidney and liver are hubs in the network (Figure 3B). The heart has the most fibrotic disease-RNA-related datasets, with 209 different types of datasets (Figure 3B). Moreover, it is the miRNAs in fibrotic diseases that have been most studied (Figure 3C).
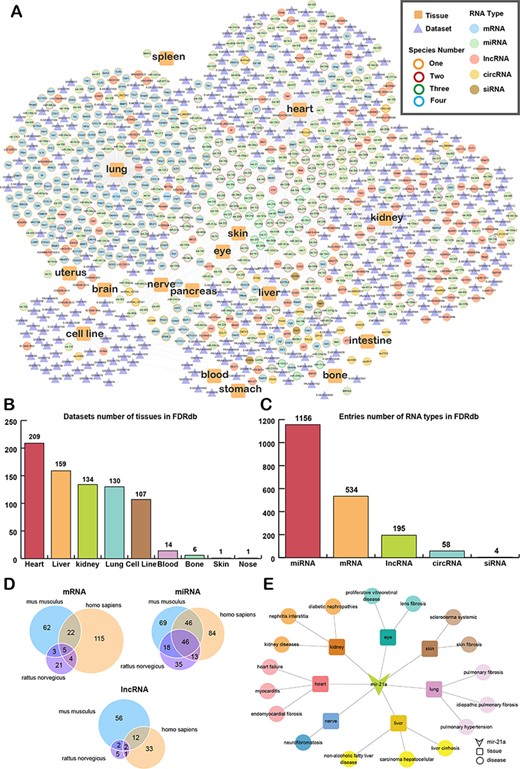
The landscape of data in the FDRdb. (A) The network of associations between fibrotic diseases and RNAs/datasets in different tissues. (B) The statistics of tissues with fibrotic datasets in the FDRdb. (C) The statistics of fibrotic RNAs in the FDRdb. (D) The Venn diagram showing the RNAs common to different species. (E) The RNA-tissue-disease sub-network of mir-21a.
We also made integrative analyses of the fibrotic RNAs (mRNA, miRNA and lncRNA) in M. musculus, H. sapiens and R. norvegicus (Figure 3D) and found that there were common fibrotic RNAs in different species. There was a large proportion of common fibrotic miRNAs among the three species, which suggested that these miRNAs were highly conservative and might have similar biological effects on fibrotic diseases. For example, mir-21a was correlated most to the tissues and fibrotic diseases of the three species (Figure 3E). In liver fibrosis, mir-21a enhanced the extracellular signal-regulated kinase 1 pathway and epithelial–mesenchymal transition processing in H. sapiens and R. norvegicus (29, 30). Mir-21a also elevated the amount of proinflammatory cytokines in dominant-negative transforming growth factor-β receptor type II mice with liver fibrosis (31). These findings suggest that mir-21a might play a key role in fibrotic diseases.
Discussion
Fibrosis, also known as fibrotic scarring, is a pathological wound healing in which connective tissue replaces normal parenchymal tissue to the extent that it goes unchecked. This leads to considerable tissue remodeling and permanent scar tissue formation. In recent years, studies have attempted to uncover the molecular regulatory mechanisms of fibrotic diseases (32), and, with the growing advancements in second-generation sequencing and other technologies, increasingly more relevant datasets have been produced. However, the majority of the relevant information is scattered in separate pieces of literature or databases, which is not conducive to helping researchers to make new summaries and discoveries of published research results. Moreover, many studies have shown a strong link between COVID-19 and fibrotic diseases. Mahmud et al. found 65 co-differential mRNAs in COVID-19 patients with chronic obstructive pulmonary disease and IPF and identified potential drug targets based on these differentially expressed mRNAs (33). Six of these genes (claudin 5, C-X-C motif chemokine ligand 12, major histocompatibility complex, class I, B, CD28 molecule, interleukin 6 and C-X-C motif chemokine receptor 4) were verified experimentally and included in our database, FDRdb. This suggests that the FDRdb can promote the exploration of associations between fibrotic diseases and COVID-19.
Currently, many RNA-disease databases have been developed, such as LncRNADisease 2.0, Circad, circRNADisease and miR2Disease. However, to the best of our knowledge, none of these databases have manually collected experimentally supported data concerning the RNAs associated with fibrotic diseases. For example, miR2Disease contains 274 relationships between 163 miRNAs and eight fibrotic diseases and Circad has 102 pieces of association data concerning 100 circRNAs and 10 fibrotic diseases. The current version of the FDRdb contains 1947 entries about associations between 912 RNAs and 92 fibrotic diseases, which contains nearly 7 and 20 times the fibrotic disease–associated RNA entries than miR2Disease and Circad, respectively. Moreover, these existing databases always focus on a single type of RNA, such as miRNA or lncRNA, and most of the data in these databases are about humans. The above-mentioned databases do not include the corresponding high-throughput datasets, download links and analysis tools, which are the highlights of the FDRdb.
In addition to containing more entries, the FDRdb has the following advantages over the other existing databases. First, the FDRdb provides information on more types of RNA in various species. Second, the FDRdb records a large number of experimentally verified data, which increases the authenticity and reliability of the association. Third, the FDRdb also collects various fibrotic disease–related datasets from different databases, which will make it easier for researchers to download and use. For example, by querying ‘miR-200a’, the FDRdb provides the miR-200a expression pattern in three different diseases among three species. These result entries are supported by experimental evidence or bioinformatics prediction. In addition, the FDRdb provides a function description for each result entry. For example, in the ‘details’ of the ‘miR-200a’-‘idiopathic pulmonary fibrosis’-‘Mus musculus’ entry, the FDRdb shows that the expression level of mir-200a is reduced in patients with IPF and suggests that restoring the miR-200a expression in the lung may be a novel therapeutic strategy for IPF. Furthermore, this entry is supported by western blot and qRT-PCR (34). Finally, we incorporated analysis tools in the FDRdb to analyze and visualize the fibrosis datasets. The information provided by the FDRdb may be helpful for biologists and pathologists to reveal new mechanisms of operation for fibrotic diseases.
In summary, the FDRdb not only focuses on the association between RNAs and fibrotic diseases but also collects datasets of fibrotic diseases for researchers to download and use. Meanwhile, the FDRdb provides a convenient web tool to analyze the expression profiles of fibrotic diseases.
In the future, we will continue to manually collect the newly validated fibrotic disease–RNA relationships and update the database semi-annually. Additionally, more RNA functional features and information will be added, such as the information on RNA alternative splicing, and we will provide prediction tools for fibrotic disease–RNA associations. In addition, we will incorporate tools in the FDRdb to analyze and visualize the fibrosis datasets.
Data availability
All data of FDRdb can be obtained in http://www.medsysbio.org/FDRdb.
Author contributions
C.W. contributed to software, data curation, visualization and writing—original draft. T.C. contributed to data curation and writing—review and editing. Y.M., X.L., K.X. and L.A. contributed to data curation. Y.G. contributed to project administration, writing—review and editing, and funding acquisition. X.F. contributed to project administration and writing—review and editing. H.L. contributed to project administration, funding acquisition, writing—review and editing, and supervision.
Funding
National Natural Science Foundation of China (grant number 91949109 and 32270710).
Conflict of interest
None declared.
References
Author notes
contributed equally to this work.



