-
PDF
- Split View
-
Views
-
Cite
Cite
William R Rolling, Douglas Senalik, Massimo Iorizzo, Shelby Ellison, Allen Van Deynze, Philipp W Simon, CarrotOmics: a genetics and comparative genomics database for carrot (Daucus carota), Database, Volume 2022, 2022, baac079, https://doi.org/10.1093/database/baac079
Close - Share Icon Share
Abstract
CarrotOmics (https://carrotomics.org/) is a comprehensive database for carrot (Daucus carota L.) breeding and research. CarrotOmics was developed using resources available at the MainLab Bioinformatics core (https://www.bioinfo.wsu.edu/) and is implemented using Tripal with Drupal modules. The database delivers access to download or visualize the carrot reference genome with gene predictions, gene annotations and sequence assembly. Other genomic resources include information for 11 224 genetic markers from 73 linkage maps or genotyping-by-sequencing and descriptions of 371 mapped loci. There are records for 1601 Apiales species (or subspecies) and descriptions of 9408 accessions from 11 germplasm collections representing more than 600 of these species. Additionally, 204 Apiales species have phenotypic information, totaling 28 517 observations from 10 041 biological samples. Resources on CarrotOmics are freely available, search functions are provided to find data of interest and video tutorials are available to describe the search functions and genomic tools. CarrotOmics is a timely resource for the Apiaceae research community and for carrot geneticists developing improved cultivars with novel traits addressing challenges including an expanding acreage in tropical climates, an evolving consumer interested in sustainably grown vegetables and a dynamic environment due to climate change. Data from CarrotOmics can be applied in genomic-assisted selection and genetic research to improve basic research and carrot breeding efficiency.
Introduction
Well-curated ‘omic’ data in an openly accessible database are likely the best way for plant scientists to disseminate information for research and breeding programs (1). This is important because genomic data are not biologically meaningful without analyses and interpretation, and the raw data are not easily provided through traditional publications. Plant genomic databases have been instrumental for improved gene annotation and understanding of genome structure. This is because they make community engagement straightforward, and a broad group of researchers can access the data to manually curate computer predictions using experimental evidence for gene function (2–4). Arabidopsis is the gold standard for plant genomics, and with community engagement, over 94% of genes have a predicted function and 48% of genes have some experimental evidence for predicted function (5).
Plant genomes harbor tremendous diversity in size, structure and evolutionary history (5). As such, having genetically related species with high-quality genome assemblies can be helpful for gene prediction when assembling a new genome. Currently, many economically important families have high-quality reference genomes and databases available, including maize (6) and rice (7) for Poaceae and Soybean (8) and Medicago (9) for Fabaceae. Rice is a great example of how a high-quality reference genome can be leveraged in the assembly of additional related genomes, and more than 100 Poaceae genomes have been sequenced following the rice genome (5).
Databases are useful for more than just reference genome analysis. For example, databases have been used to combine results of genetic mapping studies and produce more accurate estimations of the genetic positions of quantitative trait loci (QTL) using Meta-QTL analysis. This type of analysis is more practical when mining results from a database as highlighted in examples from cotton (10), corn (11) and soybean (12). Databases are also useful to generate hypotheses about which genes underlie important traits. For example, the Genome Database for Rosaceae (13) has been used as a source of data to study important gene families such as MADS-box in plum (14) and NF-Y genes in peach (15). Similarly, the CottonGen genome database (16) has been used to identify positional candidate genes following QTL mapping (17) or provide primer sequences for transcriptomic analyses of positional candidate genes (18).
In this manuscript, we describe the CarrotOmics (https://carrotomics.org/) database, which was built to disseminate carrot genomic information and encourage community involvement in carrot genetic research, breeding and germplasm study. A whole-genome sequence for carrot (Daucus carota L.) was produced in 2014 (19), and these sequences were initially available for download via website, but no broad functioning, publicly available genome database has been maintained to organize carrot genomics data. CarrotOmics was developed to provide unrestricted access to carrot genomic resources such as the high-quality reference genome sequence, gene prediction, annotation and expression data from an orange-colored, Nantes-type, doubled-haploid carrot (20). CarrotOmics also stores data from genetic mapping, phenomic, functional genomics and taxonomic studies.
Carrot is one of many economically important plant Apiaceae species, and the availability of the carrot genome at CarrotOmics will make future Apiaceae reference genome assembly and comparative genomic analyses more straightforward. For example, celery is closely related to carrot, and the celery reference genome was assembled using identity and coverage thresholds determined based on methods from the carrot genome assembly. The celery reference genome also used the carrot genome to consider synteny, genome size and gene content (21). A genome sequence is also available for the Apiales crops coriander (22) and ginseng (23), and the combination of data from these genomes can provide a basis for future comparative genomic studies.
CarrotOmics is also intended as a resource for carrot breeders. Carrot germplasm improvement is of interest because carrots are highly nutritious (24) and will help contribute to nutritional security in the 21st century. In addition to the fresh vegetable market, carrots are also the raw material for valuable commodities including juices, natural food coloring and baby food. The nutritional and monetary value has encouraged additional carrot cultivation, and production has increased 6-fold from 1970 to 2018, placing carrots among the 10 most produced vegetables, globally (25). This increasing production trend is likely to continue as East Asian countries are growing more carrots (24), and new production regions are being established in subtropical regions (25), both of which will require novel cultivars adapted for those agroecosystems.
Many of the new production regions have challenging climates and diverse agronomic practices for carrot production. New cultivars adapted for these agroecosystems are required. However, with each generation of seed production and selection requiring a full year, the current carrot breeding cycles take approximately 7 years to develop new lines (26). Carrot breeding programs can increase breeding efficiency or genetic gain in a breeding cycle through the application of genomic-assisted selection. Data curated on CarrotOmics can be helpful to identify variants for marker-assisted selection or produce genomic selection models.
With these goals in mind, carrot researchers have been collecting large genotypic and phenotypic datasets for many diverse accessions. One application has been to study the consequences of domestication and the population structure in carrot landraces and cultivars. These studies have found gene flow from wild populations to cultivars (27); genetic clustering of more related groups including wild, East Asian, Central Asian, Western (Europe and the Americas) and modern Western germplasm (28) and even genetic differences between market class of carrots (29).
In addition to being genotyped, many accessions have been phenotyped to identify genetic markers linked to economically important traits via genome-wide association studies (GWAS). For example, a novel carotenoid accumulation locus was identified by scoring for the presence or absence of orange pigmentation in more than 600 accessions (27). GWAS results complement mapping completed in more than 90 biparental populations, identifying 354 QTL (Spring 2022) for traits such as carotenoid accumulation (30), anthocyanin accumulation (31) and nematode resistance (32). Recent mapping studies have been improved because markers can be anchored to physical positions and positional candidate genes identified with the availability of the high-quality reference genome (20, 30–32).
The examples outlined here and many more studies have provided a large base for genomic-assisted breeding. CarrotOmics was developed to make that data and the reference genome available to breeders and the Apiaceae research community.
Database summary
Genome sequences available for download
Carrot reference genome—https://carrotomics.org/bio_data/290811
Other Apiaceae and organellar genomes—https://carrotomics.org/node/175
Transcriptomic data—https://carrotomics.org/node/94
A comprehensive list of Apiaceae germplasm accessions stored by germplasm centers
Germplasm search—https://carrotomics.org/search/germplasm
Descriptions, images and phenotypic evaluations of carrot germplasm
Phenotype search—https://carrotomics.org/search/qualitative_traits
Genomic & data search tools
Published carrot linkage maps and QTL.
User help guides
User manual—https://carrotomics.org/userManual
Contact information—https://carrotomics.org/contact
CarrotOmics framework
CarrotOmics is implemented using Tripal (33), a suite of Drupal modules for creating biological websites. Internal storage uses the GMOD/Chado v1.3 schema (34). CarrotOmics supports Tripal v3 RESTful web services (https://carrotomics.org/web-services/). This interface allows access to all publicly available data programmatically and allows communication with other Tripal sites. CarrotOmics also includes a BrAPI Tripal module providing a standardized application programming interface. Breeding API (BrAPI) (https://brapi.org/) helps different plant breeding databases communicate inputs, outputs, structure and how data are passed from the database website (35).
The CarrotOmics database was developed using Tripal Core modules and custom modules developed by the MainLab Bioinformatics Crop Genomics core (https://www.bioinfo.wsu.edu/). Custom modules from the MainLab include the Tripal MegaSearch (36), Tripal Map (37), Chado display, Chado search and MCL (38) and an Expression Module (39). These custom modules may be downloaded from https://gitlab.com/mainlabwsu. Custom features implemented for CarrotOmics (https://github.com/carrotomics/) are housed in a custom module or in modifications to existing modules (i.e. chado_search, mcl).
Other crop databases that were developed using MainLab resources include the Rosaceae (13), Cotton (16), Citrus (40), Pulse (41) and Vaccinium (https://www.vaccinium.org/). The homepages of these databases serve as hubs to access all data and tools.
The CarrotOmics homepage (Figure 1) includes community-specific news (Figure 1A), CarrotOmics updates and a Quick Start with direct links to data and tools (Figure 1B). Pull-down menus at the top of the homepage provide access to all database tools and data (Figure 1C). Additional news, database background information and upcoming or recent updates can be found in the ‘General’ pull-down menu (Figure 1D). Questions about the database or tools can be found in ‘User Manual’ or ‘Contact’ pages and in the ‘Help’ pull-down menu (Figure 1D).
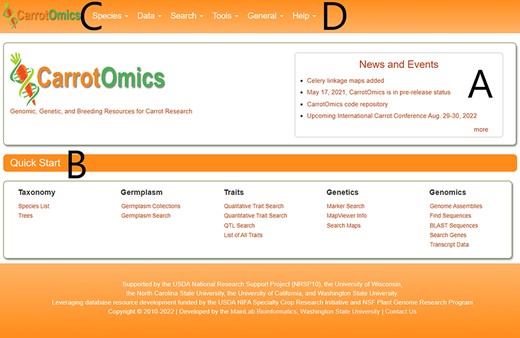
The CarrotOmics homepage includes (A) a news and events banner. (B) a Quick Start menu to directly access tools or data. (C) Pull-down menus to access all parts of the database. (D) A ‘Help’ pull-down menu to access user manuals and tutorials.
Genome & transcript data
A Nantes-type doubled-haploid carrot (Table 1) was sequenced to produce a high-quality reference genome, with the assembly accounting for >90% of the estimated genome size and 32 113 predicted genes (20). The full genome sequence, mRNA, exon, coding sequence (CDS), protein sequences and predicted gene function are available for download (Table 1) from either the ‘Genome Assemblies’ or ‘Data Download’ links in the ‘Data’ pull-down menu. Additional genomic features are available from the ‘Data Download’ (Table 1) including the predicted pathogen receptor genes (42) and a catalog of single nucleotide polymorphisms (SNPs), mobile elements, tandem repeats and gaps in the assembly.
| Description . | Search location . |
|---|---|
| Reference genome files | |
| Genome sequencea | https://carrotomics.org/bio_data/290811 |
| Gene annotations | https://carrotomics.org/file/188230 |
| Coding sequence | https://carrotomics.org/file/188228 |
| Polypeptide sequence | https://carrotomics.org/file/188229 |
| Results of reference genome analyses | |
| Tandem repeats | https://carrotomics.org/Analysis/177694 |
| Genetic variants | https://carrotomics.org/Analysis/177695 |
| Gene predictions | https://carrotomics.org/Analysis/79607 |
| Pathogen receptor genes | https://carrotomics.org/Analysis/177878 |
| Gaps & gap fill | https://carrotomics.org/Analysis/177692 |
| https://carrotomics.org/Analysis/177698 | |
| Transcriptomes | |
| Iorizzo et al. 2011b | https://carrotomics.org/bio_data/169050 |
| DCAR V1.0 gene predictionc | https://carrotomics.org/bio_data/79607 |
| NCBI | https://carrotomics.org/bio_data/70510 |
| Description . | Search location . |
|---|---|
| Reference genome files | |
| Genome sequencea | https://carrotomics.org/bio_data/290811 |
| Gene annotations | https://carrotomics.org/file/188230 |
| Coding sequence | https://carrotomics.org/file/188228 |
| Polypeptide sequence | https://carrotomics.org/file/188229 |
| Results of reference genome analyses | |
| Tandem repeats | https://carrotomics.org/Analysis/177694 |
| Genetic variants | https://carrotomics.org/Analysis/177695 |
| Gene predictions | https://carrotomics.org/Analysis/79607 |
| Pathogen receptor genes | https://carrotomics.org/Analysis/177878 |
| Gaps & gap fill | https://carrotomics.org/Analysis/177692 |
| https://carrotomics.org/Analysis/177698 | |
| Transcriptomes | |
| Iorizzo et al. 2011b | https://carrotomics.org/bio_data/169050 |
| DCAR V1.0 gene predictionc | https://carrotomics.org/bio_data/79607 |
| NCBI | https://carrotomics.org/bio_data/70510 |
fasta or gff3 files containing data from the reference genome.
Name of study on transcriptomics page (https://carrotomics.org/node/94).
Transcriptomes assembled to the reference genome currently stored at CarrotOmics.
| Description . | Search location . |
|---|---|
| Reference genome files | |
| Genome sequencea | https://carrotomics.org/bio_data/290811 |
| Gene annotations | https://carrotomics.org/file/188230 |
| Coding sequence | https://carrotomics.org/file/188228 |
| Polypeptide sequence | https://carrotomics.org/file/188229 |
| Results of reference genome analyses | |
| Tandem repeats | https://carrotomics.org/Analysis/177694 |
| Genetic variants | https://carrotomics.org/Analysis/177695 |
| Gene predictions | https://carrotomics.org/Analysis/79607 |
| Pathogen receptor genes | https://carrotomics.org/Analysis/177878 |
| Gaps & gap fill | https://carrotomics.org/Analysis/177692 |
| https://carrotomics.org/Analysis/177698 | |
| Transcriptomes | |
| Iorizzo et al. 2011b | https://carrotomics.org/bio_data/169050 |
| DCAR V1.0 gene predictionc | https://carrotomics.org/bio_data/79607 |
| NCBI | https://carrotomics.org/bio_data/70510 |
| Description . | Search location . |
|---|---|
| Reference genome files | |
| Genome sequencea | https://carrotomics.org/bio_data/290811 |
| Gene annotations | https://carrotomics.org/file/188230 |
| Coding sequence | https://carrotomics.org/file/188228 |
| Polypeptide sequence | https://carrotomics.org/file/188229 |
| Results of reference genome analyses | |
| Tandem repeats | https://carrotomics.org/Analysis/177694 |
| Genetic variants | https://carrotomics.org/Analysis/177695 |
| Gene predictions | https://carrotomics.org/Analysis/79607 |
| Pathogen receptor genes | https://carrotomics.org/Analysis/177878 |
| Gaps & gap fill | https://carrotomics.org/Analysis/177692 |
| https://carrotomics.org/Analysis/177698 | |
| Transcriptomes | |
| Iorizzo et al. 2011b | https://carrotomics.org/bio_data/169050 |
| DCAR V1.0 gene predictionc | https://carrotomics.org/bio_data/79607 |
| NCBI | https://carrotomics.org/bio_data/70510 |
fasta or gff3 files containing data from the reference genome.
Name of study on transcriptomics page (https://carrotomics.org/node/94).
Transcriptomes assembled to the reference genome currently stored at CarrotOmics.
Gene expression data are also available via the ‘Transcripts and Expression Data’ link (Table 1) in the ‘Data’ pull-down menu. Carrot researchers can provide results of transcriptomic studies, and there are currently (Spring 2022) data available from four studies (20, 43–45). The transcriptomic data can be searched by gene or transcript ID using the ‘Gene and Transcript Search’ tool found in the ‘Search’ pull-down menu.
Germplasm & phenotypic data
Germplasm, genomic and phenotypic data are also deposited on CarrotOmics. A summary is in the ‘Data’ pull-down menu (Figure 1C), ‘Data Overview’ link. Currently (Spring 2022), there are 9408 germplasm accession records from 1601 species, 73 genetic maps, 11 224 genetic markers, 371 mapped loci, 28 517 phenotypic measures and 32 662 publications (Table 2).
| Description . | Records . | Search tool location . |
|---|---|---|
| Markers | 11 224 | https://carrotomics.org/search/markers |
| Genetic maps | 73 | https://carrotomics.org/search/featuremap/list |
| QTL | 354 | https://carrotomics.org/search/qtl |
| Publication | 32 662 | https://carrotomics.org/find/publications |
| Germplasm records | 9408 | https://carrotomics.org/search/germplasm |
| Phenotypes | 28 517 | https://carrotomics.org/search/qualitative_traits |
| Phylogenetic trees | 60 | https://carrotomics.org/node/73 |
| Description . | Records . | Search tool location . |
|---|---|---|
| Markers | 11 224 | https://carrotomics.org/search/markers |
| Genetic maps | 73 | https://carrotomics.org/search/featuremap/list |
| QTL | 354 | https://carrotomics.org/search/qtl |
| Publication | 32 662 | https://carrotomics.org/find/publications |
| Germplasm records | 9408 | https://carrotomics.org/search/germplasm |
| Phenotypes | 28 517 | https://carrotomics.org/search/qualitative_traits |
| Phylogenetic trees | 60 | https://carrotomics.org/node/73 |
| Description . | Records . | Search tool location . |
|---|---|---|
| Markers | 11 224 | https://carrotomics.org/search/markers |
| Genetic maps | 73 | https://carrotomics.org/search/featuremap/list |
| QTL | 354 | https://carrotomics.org/search/qtl |
| Publication | 32 662 | https://carrotomics.org/find/publications |
| Germplasm records | 9408 | https://carrotomics.org/search/germplasm |
| Phenotypes | 28 517 | https://carrotomics.org/search/qualitative_traits |
| Phylogenetic trees | 60 | https://carrotomics.org/node/73 |
| Description . | Records . | Search tool location . |
|---|---|---|
| Markers | 11 224 | https://carrotomics.org/search/markers |
| Genetic maps | 73 | https://carrotomics.org/search/featuremap/list |
| QTL | 354 | https://carrotomics.org/search/qtl |
| Publication | 32 662 | https://carrotomics.org/find/publications |
| Germplasm records | 9408 | https://carrotomics.org/search/germplasm |
| Phenotypes | 28 517 | https://carrotomics.org/search/qualitative_traits |
| Phylogenetic trees | 60 | https://carrotomics.org/node/73 |
Germplasm records include data for 881 accessions of cultivated carrot (D. carota subsp. sativus or var. sativus). Although carrot is the focal species, CarrotOmics also serves as a data resource for other plant species from the Apiales order. Currently, seven families are represented: (i) Apiaceae, (ii) Araliaceae, (iii) Griseliniaceae, (iv) Myodocarpaceae, (v) Pennantiaceae, (vi) Pittosporaceae and (vii) Torricelliaceae. All families can be searched in the ‘Species’ pull-down menu, ‘Organism Summary’ link (Table 2).
Species information also includes a National Center for Biotechnology Information (NCBI) Taxonomic species tree found in the ‘CarrotOmics Taxonomy Tree’ link and phylogenetic relationships within the Daucus genus found in the ‘Phylogenetic Trees’ link (Table 2). The combination of wild carrot, carrot landraces and crop wild relatives provides germplasm accessions to screen for novel traits to improve abiotic stress tolerance (46) and disease resistance traits or mine data from previously screened accessions [e.g. see Alternaria resistance scores (47)].
Those accessions that have been phenotyped can be queried in the ‘Quantitative’ or ‘Qualitative Trait Search’ found in the ‘Search’ pull-down menu (Table 2). A total of 53 qualitative measurements and 70 quantitative measurements from the United States Department of Agriculture (USDA)-National Plant Germplasm System are stored at CarrotOmics. Three additional quantitative traits including Alternaria resistance (47), canopy height (48) and flavor scores from in-house germplasm screening projects are also stored on CarrotOmics.
Phenotypic data will continue to be added and updated by database curators, and individual researchers may contribute peer-reviewed or biologically replicated preprint data via the ‘Data Submission’ link found in the ‘Data’ pull-down menu.
Genetic mapping & marker data
There are 73 genetic maps currently stored on CarrotOmics, and users will be able to submit the results of their (future) mapping studies via the ‘Data Submission’ page. Existing genetic maps are searchable at the ‘Map Search’ link found in the ‘Search’ pull-down menu (Table 2). Results of map searches are organized into a table with a link to the map and descriptions of the mapping population including parental accessions, mapping population size and generation. A related function in the ‘Search’ pull-down menu is ‘Marker Search’ (Table 2). Markers can be searched by (sub)species, marker type, mapping population, linkage group and, if available, the trait that the marker is associated with from the mapping study or association analysis. Published marker-trait associations can also be found with the ‘QTL and MTL Search’ (Table 2).
Tools & references
There are many search tools for specific types of data on CarrotOmics. However, there is also the ‘MegaSearch’ (36) tool in the ‘Search’ pull-down menu. ‘MegaSearch’ is a broad functioning search module that can be used to query the entirety of the database. Users can select data types of interest, filter the data, customize fields and download data from a single interface.
There are four analysis tools currently available via ‘Tools’ pull-down menu including (i) ‘BLAST+’, (ii) ‘JBrowse’, (iii) ‘MapViewer’ and (iv) ‘Expression Heatmap’. The ‘BLAST+’ tool (49) links to blastn, blastx, tblastn and blastp analyses with options to select a database and specify advanced BLAST options.
A useful tool to visualize genetic variants and associated linkage maps is the ‘MapViewer’ tool, which includes a video tutorial (Figure 2). The tool can be used to visualize and compare genetic maps from different populations. For example, the ‘Y2’ locus related to carotenoid accumulation (45) was mapped to a region on linkage group 7 (corresponds to Chr. 07) (Figure 2B). Information about this QTL can be accessed directly in MapViewer (Figure 2C) or through a link to a page about the QTL (Figure 2D). The physical positions of markers flanking the QTL can also be accessed through links on MapViewer (Figure 2E).
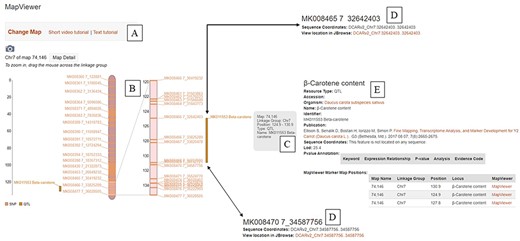
The ‘MapViewer’ tool available at CarrotOmics. (A) Links to video and text tutorials on use of ‘MapViewer’. (B) Linkage map display from Y2 QTL mapping study. (C) Information about QTL displayed on interactive ‘MapViewer’ plot. (D) Physical positions for markers flanking Y2 locus. (E) Information about the study used to create the linkage map.
JBrowse can be used to browse genome assemblies (50) (Figure 3). The carrot genome ‘JBrowse’ currently contains 17 tracks to visualize sequences, gaps and gene annotations from NCBI and the reference genome publication (20). There are tracks for mRNA data, including those used in the genome annotation (20) and from a more recent transcriptome study of carotenoid synthesis (44). Five tracks are available for notable genome features including transposons and repetitive elements. Another set of five tracks represents genetic variants used in genetic mapping or identified in the reference genome study (20).
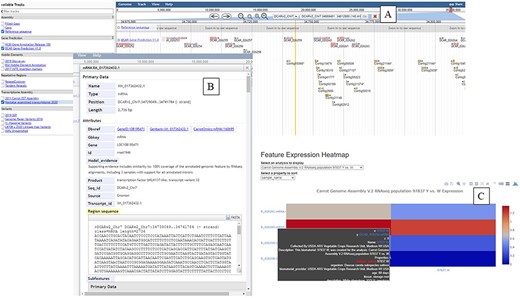
The ‘JBrowse’ tool available at CarrotOmics. (A) ‘JBrowse’ console with gene annotation tracks displayed. (B) Information about selected gene. (C) Heatmap display from transcriptome studies available from ‘Expression Heatmap’ tool.
The final tool is an ‘Expression Heatmap’ to visualize transcript expression data. Specific carrot transcripts of interest can be searched based on identifier or annotation (Figure 3C). Once transcripts of interest are selected, a heatmap displaying relative expression levels is displayed and available for export. These plots can be customized by selecting a specific transcriptome analysis or type of tissue and biomaterial used.
Conclusions and future directions
CarrotOmics was developed to serve as a hub for carrot breeding and Apiaceae genomics research. The initial release of CarrotOmics includes a high-quality reference genome, a plethora of genetic maps, germplasm descriptions and gene expression data. The database will be updated as new or improved genome assemblies are produced, more genotypic and phenotypic data become available and additional transcriptomic studies are completed.
We anticipate this database will be maintained in a similar manner as those other databases hosted by the MainLab, including CottonGen (16), Genome Database for Rosaceae (13), Citrus Genome Database (40), Pulse Crop Database (41) and Genome Database for Vaccinium (https://www.vaccinium.org/). Specifically, the USDA carrot genomic and breeding lab has been supported for over 50 years, and carrots continue to be an important vegetable crop with USDA-Vegetable Crop Research Unit support, external USDA-National Institute of Food and Agriculture (NIFA) grants and cooperative agreements with industry partners. We anticipate CarrotOmics will become an integral part of the USDA-Carrot research crop program, specifically with the addition of the breeding management system (51), which is BrAPI complaint. This function will be particularly useful as the USDA moves to digital agricultural breeding platforms (i.e. Breeding Insights).
CarrotOmics provides novel data and functionality not currently available in other Apiaceae databases including descriptions and phenotypic data from the USDA-Germplasm Resources Information Network (GRIN) germplasm collections for Apiales species, Apiales publication search tools and phylogenetic analyses. CarrotOmics also describes the results of QTL mapping studies including information about the physical positions of QTL, the mapping populations and nearby markers. Researchers from other Apiaceae species are invited to upload data to CarrotOmics and use tools not yet available on other databases including MapViewer, Expression Heatmaps and MegaSearch. We have added google analytics to track page visits and download counts to identify which areas of the database are most used and put database resources toward developing those tools.
In time, we also hope to provide links to data analysis pipelines and tutorials on how to use the deposited data for genomic studies such as genomic prediction, genome-wide association analyses, Mendelian and biparental mapping and calculation of genetic values of breeding lines. CarrotOmics will also house nontechnical resources for outreach events and educational activities, cultivars released, seed availability and video demonstrations for the culinary uses of carrot, as they become available. We hope these resources as well as the genomic tools will support an active global community of breeders, molecular biologists, growers and horticulturists.
Acknowledgements
The authors would like to specifically thank Dorrie Main, Heidi Hough, Taein Lee, Katheryn Buble and Chun-Huai Cheng for assistance in setting up the database and the entire Tripal development community for their contributions to Tripal Core and Tripal modules. The authors thank all those who have contributed genomic and phenotypic data stored at CarrotOmics.
Funding
This work was supported by the United States Department of Agriculture National Institute of Food and Agriculture Specialty Crop Research Initiative project 2016–01957. Partial salary support for WR was provided by the United States Department of Agriculture National Institute of Food and Agriculture project 5090-21000-069-061-I.
Author contributions
D.S. formatted and uploaded data to the CarrotOmics database and developed the CarrotOmics Tripal module. W.R.R. assisted with database formatting and wrote the original version of the manuscript. M.I., S.E., A.V.D., D.S. and P.W.S. conceived the project and provided technical suggestions for the database. All authors read, revised and approved the final version of this manuscript.
Conflict of interest
The authors declare no competing interest.
Data availability
All data described in this manuscript is available at https://www.carrotomics.org/.



