-
PDF
- Split View
-
Views
-
Cite
Cite
Anfu Tang, Louise Deléger, Robert Bossy, Pierre Zweigenbaum, Claire Nédellec, Do syntactic trees enhance Bidirectional Encoder Representations from Transformers (BERT) models for chemical–drug relation extraction?, Database, Volume 2022, 2022, baac070, https://doi.org/10.1093/database/baac070
Close - Share Icon Share
Abstract
Collecting relations between chemicals and drugs is crucial in biomedical research. The pre-trained transformer model, e.g. Bidirectional Encoder Representations from Transformers (BERT), is shown to have limitations on biomedical texts; more specifically, the lack of annotated data makes relation extraction (RE) from biomedical texts very challenging. In this paper, we hypothesize that enriching a pre-trained transformer model with syntactic information may help improve its performance on chemical–drug RE tasks. For this purpose, we propose three syntax-enhanced models based on the domain-specific BioBERT model: Chunking-Enhanced-BioBERT and Constituency-Tree-BioBERT in which constituency information is integrated and a Multi-Task-Learning framework Multi-Task-Syntactic (MTS)-BioBERT in which syntactic information is injected implicitly by adding syntax-related tasks as training objectives. Besides, we test an existing model Late-Fusion which is enhanced by syntactic dependency information and build ensemble systems combining syntax-enhanced models and non-syntax-enhanced models. Experiments are conducted on the BioCreative VII DrugProt corpus, a manually annotated corpus for the development and evaluation of RE systems. Our results reveal that syntax-enhanced models in general degrade the performance of BioBERT in the scenario of biomedical RE but improve the performance when the subject–object distance of candidate semantic relation is long. We also explore the impact of quality of dependency parses. [Our code is available at: https://github.com/Maple177/syntax-enhanced-RE/tree/drugprot (for only MTS-BioBERT); https://github.com/Maple177/drugprot-relation-extraction (for the rest of experiments)]
Database URLhttps://github.com/Maple177/drugprot-relation-extraction
Introduction
Relation extraction (RE) is an essential task in the domain of Natural Language Processing (NLP) and biomedical information extraction. The goal of biomedical RE is to automatically detect relations between biomedical entities in scientific texts, thus facilitating the retrieval of valuable biomedical information. Compared to manual extraction, an effective automatic RE system consumes much less time and demands fewer human resources. In most cases of RE, relations refer to pre-defined semantic relations between entities, such as relations between chemicals and genes/proteins. An example is given in Figure 1, where we can see the chemical ‘Caffeine’ linked to the protein ‘kinase’ by the ‘INHIBITOR’ relation.

An example drawn from the DrugProt data set, showing a sentence with an INHIBITOR relation between a chemical/drug entity (the subject, Caffeine) and a gene/protein entity (the object, kinase).
Over recent years, the pre-trained transformer model (1) has been ubiquitous in the domain of NLP due to its superior performance over various NLP tasks. As one of the most popular pre-trained transformer models, BERT (2) has been widely applied to RE tasks. However, vanilla BERT is found to be less effective in a domain-specific scenario due to several reasons: differences between the corpus on which BERT is pre-trained and the domain-specific corpus on which it is fine-tuned and limited amount of available data for fine-tuning. To tackle this problem, different variants of BERT targeted at specific domains have been proposed, such as BioBERT (3) and SciBERT (4).
Although these BERT variants proved to outperform vanilla BERT on domain-specific tasks, the lack of sufficient data for fine-tuning remains a problem. Therefore, we seek to enhance their performance even further. We make the hypothesis that syntactic information might not be sufficiently encoded in BERT models and that injecting syntax into BERT may enrich the representation of the input texts and therefore improve the performance. Previous studies support this hypothesis, as in (5–7). Furthermore, existing syntactic parsers make it possible to obtain parse trees at reasonable computational cost, which facilitates the development of syntax-enhanced models. Dependency trees and constituent trees are common syntactic representations that we can obtain from syntactic parsers, as illustrated in Figure 2b.
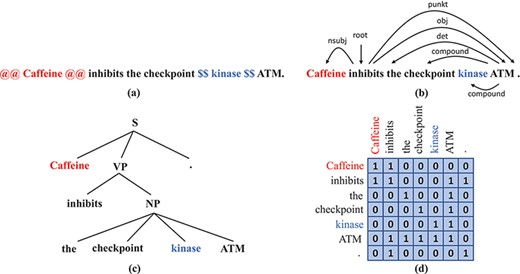
Syntax representations that we obtain from off-the-shelf parsers: (a) A sentence with the subject and the object wrapped by markers. (b) The dependency tree of the sentence in (a). (c) The constituency tree of the sentence in (a). (d) The adjacency matrix that corresponds to the dependency tree in (b).
Although some studies report that injecting syntax into BERT models improves performance on certain NLP tasks, there is no consensus on the effect of injecting syntactic information into a pre-trained BERT model. Furthermore, there is no definitive conclusion about the extent of the syntactic information implicitly encoded in BERT or the way it is encoded. To the best of our knowledge, relevant studies can be divided into two categories. The first category includes proposals of new neural network architectures that introduce syntactic information as extra input or weights inside the neural network. The second category includes work studying how and how much syntax is already implicitly encoded inside BERT. However, among the proposed syntax-enhanced neural architectures, few researchers concentrate on RE tasks, and even fewer studies have been conducted on biomedical RE tasks. We aim to fill this gap by studying syntax-enhanced BERT models for biomedical RE. In this context, we summarize our main contributions as follows:
We propose three new syntax-enhanced neural architectures:
Chunking-Enhanced-BioBERT (CE-BioBERT), enhanced with chunking information;
Constituency-Tree-BioBERT (CT-BioBERT), enhanced with encoding complete constituency trees;
Multi-Task-Syntactic-BioBERT (MTS-BioBERT), enhanced by fine-tuning with multiple tasks including a RE task and two other syntax-related tasks that aim to recover geometric properties of dependency trees.
We test an existing syntax-enhanced Late-Fusion model that was not previously evaluated on biomedical RE tasks.
We build ensemble systems containing different combinations of syntax-enhanced models and models with no syntax.
We conducted our experiments on the DrugProt corpus from the BioCreative VII track 1 shared task. In this corpus, chemical–gene relations are annotated and we seek to automatically extract relations on the test set. Details about the corpus will be given in the Materials and methods section.
Our paper is organized as follows: in the Related work section, we present syntax-related studies on the injection of syntactic information into BERT models and the capability of BERT models to encode syntactic information. Then we give details about the methods proposed and the DrugProt corpus in the Materials and methods section. In the Results section, we present the experimental setup and scores achieved by each model on the DrugProt corpus. At last we discuss results and give a conclusion about our work.
Related work
Due to their superior performance on various NLP benchmarks, pre-trained transformer models such as BERT (2) have become the basis of most recent NLP approaches, including RE approaches. Conventionally, an RE task is treated as a special kind of text classification problem as in (4, 7). A common pipeline is to first initialize a BERT model with pre-trained weights and then fine-tune the model on the data set of interest. Our work employs this workflow.
Domain-specific BERT variants
Domain-specific models have been developed to improve the performance of BERT models on domain-specific tasks. The principle behind these BERT variants is either to continue or to start from scratch the pre-training phase on a domain-specific corpus. For example, the pre-training corpus for BioBERT is enriched with PubMed abstracts and PubMed Central (PMC) full-text articles, and BioBERT is initialized with weights from vanilla BERT. Similarly, the pre-training corpus of SciBERT is enriched with scientific papers collected from Semantic Scholar: 18% are papers from the computer science domain and 82% are from the broad biomedical domain. We test both BioBERT and SciBERT in our experiments, but for syntax-enhanced models, we use only BioBERT as the base model. Notice that there exist different versions of BioBERT and SciBERT; in our experiments, we use always the BioBERT v1.1 as described in (3), which is pre-trained on English Wikipedia, BookCorpus and PubMed Abstracts using the same wordpiece vocabulary as vanilla BERT; four versions of SciBERT are provided in (4), and we always use the version pre-trained from vanilla BERT checkpoints with uncased newly constructed wordpiece vocabulary SciVocab.
Probing pre-trained BERT models
To shed light on the superior performance of BERT models and identify potential directions for further improvement, previous studies have looked into the linguistic properties of the representations learned by these models. In particular, there has been a focus on researching whether syntactic information is implicitly encoded in BERT. Probing tasks play an important role in these studies. Probing tasks, also named diagnostic classifiers, refer to simple classification tasks that are designed to test the capability of a pre-trained transformer model to encode specific types of features. Several probing tasks have thus been designed to study how BERT encodes syntactic information. If simply training a linear classifier on top of a BERT model succeeds to outperform naive baselines like non-contextualized word embeddings on predicting syntax-related labels, then it demonstrates that the pre-trained BERT model encodes some form of syntactic information. Hewitt and Manning (8) designed two probing tasks to test whether dependency trees are encoded in BERT’s embedding space. The first is a structural probe that aims to predict pairwise distances of tokens in the dependency tree given the BERT’s embedding space by calculating Euclidean distances between corresponding token embedding vectors. The second is a tree depth probe, which aims to predict the depth of each word in the dependency tree using the squared vector norm of the corresponding word embedding. Coenen et al. (9) hypothesize that the dependency graph is encoded in the weights across all attention layers of BERT. Thus, it should be possible to predict the existence of a dependency between two words or even its label, given the corresponding vector of attention weights along attention heads across all attention layers. Their experimental results show that predictions made with these vectors of attention weights are more accurate than several baseline models, such as pre-trained BERT token embeddings, or shallow contextualized embeddings at the output of a randomly initialized biLSTM layer.
Although these studies agree that BERT models do encode some sort of dependency structure, it remains unclear whether this implicit information is sufficient for fully encoding syntax trees. Still, as we will detail in the next subsection, a few recent studies have reported performance improvements on certain NLP tasks when injecting additional syntax into BERT.
Although designed to test whether syntax is encoded in BERT, probing tasks can also be used as supervised objectives to guide a neural network to learn syntax. We propose to build on these tasks for our multi-task architecture [MTS-BERT, inspired by the work of Hewitt and Manning (8)].
Neural architectures integrating syntactic information
Many neural architectures have been developed in which syntactic information is introduced explicitly or implicitly.
Explicitly injecting syntactic information consists of encoding that information partially or completely and then using the resulting syntactic representations as extra input to the neural network or embedding them inside the neural network. There exist multiple ways to encode syntactic information, among them is dependency information as organized in a syntactic dependency graph. A dependency graph is the adjacency matrix of a dependency parse tree; it gives information about whether two words are directly connected by a certain kind of dependency relation. An example of a dependency graph is given in Figure 2d. Sachan et al. (7) propose to append to BERT attention layers in which the word-to-word attention matrix is replaced by the dependency graph. Simply fine-tuning their proposed network on the TAC Relation Extraction Dataset (TACRED) RE data set (10) performed slightly better than the vanilla BERT. They also claim that dependency information helps improve the performance of BERT on RE tasks, but the improvement heavily depends on the quality of the dependency trees. Yu et al. (11) propose Dynamically Pruned Graph Convolutional Network to average the dependency graph with the attention matrix and control the information flow between words by applying a binary gate on each word. Guo et al. (12) propose Attention Guided Graph Convolutional Network (AGGCN), which consists of several stacked multi-head attention layers. Instead of totally replacing attention matrices in all layers by the dependency graph, in AGGCN the dependency graph is used to initialize the attention matrix in the first layer; in subsequent layers attention matrices are computed as in (1), i.e. attention weights are assigned to all pairs of words, including edges that do not exist in the dependency graph. Besides dependency information, some networks are developed to incorporate constituency information. Nguyen et al. (5) propose to embed nodes in the constituency tree as well as the leaves that are tokens and to design masks that make each node attend to all descendent nodes and leaves in the dependency subtree.
Implicitly introducing the syntax into BERT models does not seek to encode syntactic information in input data or inside BERT, but rather make BERT learn itself syntactic information by orienting the pre-training or fine-tuning stage using extra syntax-related tasks. Xu et al. (13) introduce a new pre-training task, Distance Prediction, whose objective is to predict distances between two words given the dependency tree of a sentence. Their results demonstrate that pre-training with extra syntax-related tasks improves model performance. Strubell et al. (6) propose a multi-task learning framework consisting of multiple stacked attention layers and train their model on four tasks containing the target Semantic Role Labelling (SRL) task and three syntactic tasks that predict part-of-speech (POS) tags and predicates, perform parsing and attend to syntactic heads, respectively. Their proposed model performs better on the SRL task than previous baselines with no syntactic information integrated. This is attributed to sharing the weights of the lower layers of the model with syntactic tasks.
It is worth noticing that some above-mentioned architectures do not involve transformer models and that most of them are not tested on biomedical RE tasks. Nevertheless, they provide possible solutions to integrate syntactic information into neural networks. The architectures that we propose in subsequent sections are inspired by these works.
Materials and methods
Methods
We investigate several approaches to inject syntax into BERT models, looking at both explicit and implicit methods and using syntactic information in the form of dependencies and constituents. We first test an existing approach not previously evaluated on biomedical RE tasks, which explicitly injects dependency-based syntactic information. We then propose two novel constituency-based explicit approaches. Finally, we introduce a multi-task method that implicitly injects syntax. The approaches are used in ensemble systems to mitigate the variance problem often encountered in neural networks.
An existing dependency-enhanced model: Late-Fusion
where |${{\rm{v}}_{\rm{i}}}{{\bf{W}}_{\bf{Q}}}$| and |${{\rm{v}}_{\rm{j}}}{{\bf{W}}_{\bf{K}}}$| refer to the corresponding query and key vector, respectively, and |${{\rm{d}}_{\rm{k}}}$| refers to the dimension of key vectors, which is a scaling term as indicated in (1).
where |${{\mathcal N}_{\rm{i}}}$| refers to the group of wordpieces that are connected to the |${\rm{i}}$|-th wordpiece in the extended dependency tree.
Although Sachan et al. (7) propose two syntax-augmented models based on syntax-GNN layers, named Late-Fusion and Joint-Fusion, we choose to test the Late-Fusion model because firstly the original paper reported an improved performance of Late-Fusion on TACRED (10) which is a benchmark for RE in the general domain, and secondly, we observed in preliminary experiments that the Joint-Fusion model heavily degraded performance on RE tasks. The Late-Fusion model consists of adding several syntax-GNN layers atop the pre-trained BERT and combining the outputs of the pre-trained BERT and the outputs of the syntax-GNN block by a Highway Gate (15) instead of the residual connection used in standard attention layers.
Two constituency-enhanced models: CE-BioBERT and CT-BioBERT
Different from the Late-Fusion model which is enhanced with dependency information, we propose to build syntax-enhanced models with constituency information, which instead of establishing pairwise relations for each word assigns words to local groups in a hierarchical way.
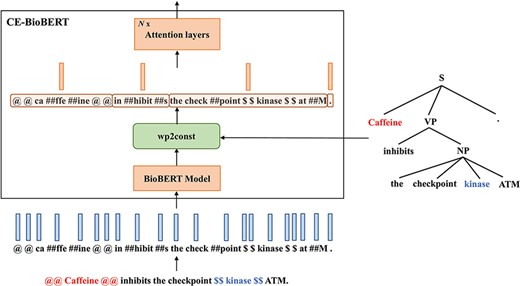
Diagram illustrating the architecture of CE-BioBERT: the wp2const block at the output of the BioBERT model groups together the wordpieces that belong to a pre-defined chunk to compute chunk embeddings.
where |${{\mathcal C}_i}$| denotes the set of wordpiece indexes that belong to the |$i$|-th constituent and |${u_i}$| the embedding of the |$i$|-th constituent.
CT-BioBERT While our first approach relied mostly on chunks, our second method, named CT-BioBERT, aims to embed complete constituent trees. Inspired by (5) in which the authors propose to embed nodes in constituency trees as well as leaves (which are tokens) and apply a subtree attention mask that makes non-leaf nodes in a constituency tree attend to all nodes and leaves in their subtree, we propose to:
Linearize the constituency tree by iterating nodes in Depth First Search (DFS) order.
Make non-leaf nodes attend to all nodes in their subtree, while all leaves will attend to each other.
The example in Figure 4 illustrates the linearization of the constituency tree that we propose. By converting a constituency tree to its DFS traversal, the resulting sequence is supposed to encode hierarchical information of the tree structure; furthermore, we keep the constituency tags of non-leaf nodes, which provides additional information about the tree. By applying subtree masks onto the attention matrix, we control the information flow between nodes so that non-leaf nodes are restricted to interact only with their descendants, and this masking also reflects the constituent hierarchy in a tree. Besides, we keep the wordpiece-to-wordpiece attention pattern, which is the same as in standard attention layers. However, we do not distinguish non-leaf nodes from leaf nodes (which are wordpieces) but treat these inserted constituency tags as independent wordpieces by adding them to the vocabulary of the wordpiece tokenizer. This operation certainly increases the difficulty of fine-tuning due to the fact that wordpiece embeddings for these newly added constituency tags are randomly initialized and the pre-trained BioBERT does not see any of them during the pre-training phase; we still keep this model in experiments however, hypothesizing that the additional information brought by constituency tags may compensate for the negative effect caused by introducing new wordpieces.
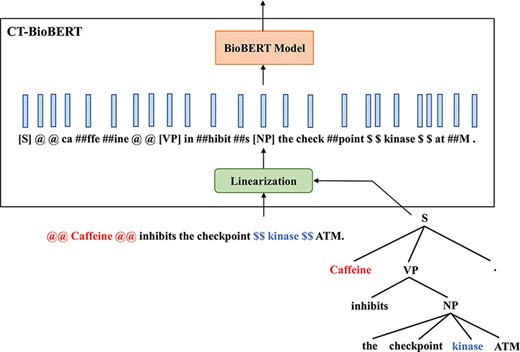
Diagram illustrating the architecture of CT-BioBERT: linearized constituency trees are fed into the BioBERT model, and each BERT layer shares the same subtree mask.
A Multi-Task-Learning framework: MTS-BioBERT
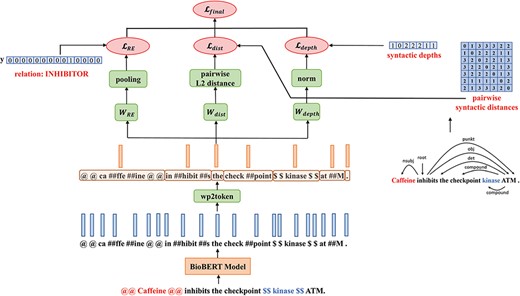
The architecture of MTS-BioBERT: Besides the relation label, for the two probing tasks, we compute pairwise syntactic distance matrices and syntactic depths from dependency trees obtained from a syntactic parser.
where |$\unicode{x03B1}$| is a weighting coefficient which controls the quantity of injected syntactic information and serves as a hyperparameter. We calculate the square root of |${\mathcal{L}_{{\rm{dist}}}} + {\mathcal{L}_{{\rm{depth}}}}$| to reduce the norm of gradients.
Ensemble systems
We adopt an ensemble strategy to evaluate our models. Ensemble methods combine predictions from multiple models to make the final prediction and are often able to outperform each individual model especially when a significant diversity exists between the models, which is the case for neural networks in the optimization scenario based on gradient-descent algorithms. In this work, for each neural architecture, we train multiple models initialized with different random seeds and combine the results of all models by simple majority voting.
Additionally, we also propose combinations of models based on different architectures, in particular models with and without syntax. We hypothesize that different types of model might make different types of predictions and errors and that combining them might boost performance and reduce errors.
Data set
The DrugProt task (16) of the BioCreative challenge provides a corpus including 14 230 PubMed abstracts and 24 526 annotated relations between chemical/drug and gene/protein entities. To prevent participants from manually annotating the test set, the organizers added dummy data (10 000 abstracts) to the manually annotated test set (750 abstracts). The statistics of the corpus are shown in Table 1. It is worth noticing that ‘chemical’ and ‘drug’ are used interchangeably in the annotation, as well as ‘gene’ and ‘protein’. An example annotation is shown in Figure 1. All chemical–gene relations are directed, and the direction is fixed from chemicals to genes. There are in total 13 classes of relations describing different interactions between genes and chemicals. We list these relation types and corresponding numbers of examples in Table 2 [Statistics in Table 2 are calculated based on our pre-processed data set. We found that statistics reported in Table 2 of the DrugProt overview article (16) were not consistent with their published corpus].
| Set . | Number of abstracts . | Number of relations . |
|---|---|---|
| Train | 3500 | 17 274 |
| Dev | 750 | 3761 |
| Test | 750 | 3491 |
| Dummy data | 10 000 | – |
| Set . | Number of abstracts . | Number of relations . |
|---|---|---|
| Train | 3500 | 17 274 |
| Dev | 750 | 3761 |
| Test | 750 | 3491 |
| Dummy data | 10 000 | – |
| Set . | Number of abstracts . | Number of relations . |
|---|---|---|
| Train | 3500 | 17 274 |
| Dev | 750 | 3761 |
| Test | 750 | 3491 |
| Dummy data | 10 000 | – |
| Set . | Number of abstracts . | Number of relations . |
|---|---|---|
| Train | 3500 | 17 274 |
| Dev | 750 | 3761 |
| Test | 750 | 3491 |
| Dummy data | 10 000 | – |
| Relation type . | Number of relations . |
|---|---|
| ACTIVATOR | 1674 |
| AGONIST | 789 |
| AGONIST-ACTIVATOR | 39 |
| AGONIST-INHIBITOR | 15 |
| ANTAGONIST | 1190 |
| DIRECT-REGULATOR | 2705 |
| INDIRECT-DOWNREGULATOR | 1661 |
| INDIRECT-UPREGULATOR | 1680 |
| INHIBITOR | 6535 |
| PART-OF | 1142 |
| PRODUCT-OF | 1078 |
| SUBSTRATE | 2497 |
| SUBSTRATE PRODUCT-OF | 27 |
| Relation type . | Number of relations . |
|---|---|
| ACTIVATOR | 1674 |
| AGONIST | 789 |
| AGONIST-ACTIVATOR | 39 |
| AGONIST-INHIBITOR | 15 |
| ANTAGONIST | 1190 |
| DIRECT-REGULATOR | 2705 |
| INDIRECT-DOWNREGULATOR | 1661 |
| INDIRECT-UPREGULATOR | 1680 |
| INHIBITOR | 6535 |
| PART-OF | 1142 |
| PRODUCT-OF | 1078 |
| SUBSTRATE | 2497 |
| SUBSTRATE PRODUCT-OF | 27 |
| Relation type . | Number of relations . |
|---|---|
| ACTIVATOR | 1674 |
| AGONIST | 789 |
| AGONIST-ACTIVATOR | 39 |
| AGONIST-INHIBITOR | 15 |
| ANTAGONIST | 1190 |
| DIRECT-REGULATOR | 2705 |
| INDIRECT-DOWNREGULATOR | 1661 |
| INDIRECT-UPREGULATOR | 1680 |
| INHIBITOR | 6535 |
| PART-OF | 1142 |
| PRODUCT-OF | 1078 |
| SUBSTRATE | 2497 |
| SUBSTRATE PRODUCT-OF | 27 |
| Relation type . | Number of relations . |
|---|---|
| ACTIVATOR | 1674 |
| AGONIST | 789 |
| AGONIST-ACTIVATOR | 39 |
| AGONIST-INHIBITOR | 15 |
| ANTAGONIST | 1190 |
| DIRECT-REGULATOR | 2705 |
| INDIRECT-DOWNREGULATOR | 1661 |
| INDIRECT-UPREGULATOR | 1680 |
| INHIBITOR | 6535 |
| PART-OF | 1142 |
| PRODUCT-OF | 1078 |
| SUBSTRATE | 2497 |
| SUBSTRATE PRODUCT-OF | 27 |
Pre-processing
In the original corpus, we find that in the training set only 3 among 17 274 (|$ \approx $|0.02%) relations are inter-sentence, while in the validation set there are no inter-sentence relations. Therefore, we choose to examine only intra-sentence relations and remove the three inter-sentence relations from the training set. We firstly segment each abstract into sentences, then enumerate all candidate pairs (subject, object) within each sentence and mark the subject and the object as presented in Figure 2. To indicate the candidate pair (subject, object), we wrap the subject in two consecutive ‘@’ characters and the object in two consecutive ‘$’ characters as in Figure 1. It is worth noticing that if the subject and the object refer to the same entity, we wrap the entity in two consecutive ‘¢’ characters.
Parsing sentences As mentioned in the Methods subsection, syntax-enhanced models require either dependency trees or constituency trees. To generate dependency trees, we choose a biomedical version of Stanza (17) trained on the GENIA treebank (18), which was collected from PubMed abstracts related to ‘transcription factors’. According to Zhang et al. (17), Stanza consistently outperformed previous state-of-the-art syntactic parsers (CoreNLP and scispaCy) on three biomedical or clinical treebanks including GENIA. Although we are unsure about the quality of syntactic parse trees that Stanza generates on biomedical texts related to gene–chemical interactions, we think the texts in GENIA are somewhat similar to those in DrugProt because both contain mentions of genes or proteins. We also use this version of Stanza for sentence segmentation. To generate constituency trees, we use the Berkeley Neural Parser (19), which is a state-of-the-art constituency parser. Subsequently, we generate concrete syntax representations for each model, for example, adjacency matrix for Late-Fusion, linearized sequences and subtree masks for CT-BioBERT, etc.
Implementation detail
Implementation of all models is based on HuggingFace’s transformer (20) and Pytorch (21). For syntax-enhanced models (CE-BioBERT, CT-BioBERT, Late-Fusion and MTS-BioBERT), we initialize parameters from BioBERT (3) version 1.1 released by HuggingFace. In cases where extra attention layers (as in CE-BioBERT or Late-Fusion) are added atop BioBERT, weights of extra layers are randomly initialized. Specifically, for CT-BioBERT, we add 24 constituency tags into the vocabulary of BioBERT’s wordpiece tokenizer, and wordpiece embeddings for newly added constituency tags are randomly initialized. We fine-tune each model with mono 32 G NVIDIA V100 Graphics processing unit (GPU) and control the fine-tuning process by early stopping, which is a common technique in deep learning to avoid overfitting by stopping when the performance on the validation set has not increased for a set number of epochs. In our experiments, we stop fine-tuning when the F1-score on the validation set has not increased for three epochs. We use the Adam (22) optimizer and keep the learning rate constant during the whole fine-tuning stage with no warm-up steps.
Experiments
In this paper, we report post-BioCreative VII experiments in which we investigate the contribution of syntax rather than aim to maximize performance. However, to give an insight into what constitutes a high performance on the DrugProt data set, we present the performance of the top performing DrugProt participants in Table 3. We see that F1-scores on this dataset range from 77.4 to 79.7 for the top six teams.
| Rank/Team ID . | Precision . | Recall . | F1-score . |
|---|---|---|---|
| #1 Team_15 | 79.6 | 79.9 | 79.7 |
| #2 Team_18 | 78.5 | 80.5 | 79.5 |
| #3 Team_13 | 79.7 | 78.2 | 78.9 |
| #4 Team_7 | 80.4 | 74.5 | 77.6 |
| #5 Team_21 (Ours) | 75.5 | 79.7 | 77.5 |
| #6 Team_3 | 77.1 | 77.7 | 77.4 |
| Rank/Team ID . | Precision . | Recall . | F1-score . |
|---|---|---|---|
| #1 Team_15 | 79.6 | 79.9 | 79.7 |
| #2 Team_18 | 78.5 | 80.5 | 79.5 |
| #3 Team_13 | 79.7 | 78.2 | 78.9 |
| #4 Team_7 | 80.4 | 74.5 | 77.6 |
| #5 Team_21 (Ours) | 75.5 | 79.7 | 77.5 |
| #6 Team_3 | 77.1 | 77.7 | 77.4 |
| Rank/Team ID . | Precision . | Recall . | F1-score . |
|---|---|---|---|
| #1 Team_15 | 79.6 | 79.9 | 79.7 |
| #2 Team_18 | 78.5 | 80.5 | 79.5 |
| #3 Team_13 | 79.7 | 78.2 | 78.9 |
| #4 Team_7 | 80.4 | 74.5 | 77.6 |
| #5 Team_21 (Ours) | 75.5 | 79.7 | 77.5 |
| #6 Team_3 | 77.1 | 77.7 | 77.4 |
| Rank/Team ID . | Precision . | Recall . | F1-score . |
|---|---|---|---|
| #1 Team_15 | 79.6 | 79.9 | 79.7 |
| #2 Team_18 | 78.5 | 80.5 | 79.5 |
| #3 Team_13 | 79.7 | 78.2 | 78.9 |
| #4 Team_7 | 80.4 | 74.5 | 77.6 |
| #5 Team_21 (Ours) | 75.5 | 79.7 | 77.5 |
| #6 Team_3 | 77.1 | 77.7 | 77.4 |
During the challenge, our best result was obtained with an ensemble system containing BioBERT and SciBERT models, which outperformed systems containing models of a same type. It demonstrates that having different types of models vote may counteract the systematic errors of each model and therefore boost the overall performance. We test ensemble systems consisting of syntax-enhanced models and non-syntax-enhanced models based on the hypothesis that these two types of model will act in a complementary way.
For post-challenge experiments, besides the syntax-enhanced models introduced in the previous section, we use three non-syntax-enhanced models as baselines:
an ensemble containing five fine-tuned BioBERT models;
an ensemble containing five fine-tuned SciBERT models;
an ensemble containing five fine-tuned modified BioBERT models in which attention layers were added atop the pre-trained BioBERT model; the number of added layers is a hyperparameter.
The third baseline is built as a contrast for the CE-BioBERT and Late-Fusion methods, as they both require the addition of extra layers to integrate syntactic information. By adding extra layers (with no syntax) into the baseline model, we ensure that the integration of syntactic information is the only difference between the second baseline and CE-BioBERT or Late-Fusion, and therefore we can make a fair comparison.
We always use the original train/dev split provided by the challenge organizers. We use the training set for training the models and monitor model performance on the validation set in order to avoid overfitting.
Hyperparameters
For each type of model, we use grid search to determine the optimal hyperparameters and instead of applying the same grid for all models, we first empirically establish a hyperparameter grid and then slightly enlarge or reduce the search space for each model according to preliminary experiments. Due to the limitation of computing resources, we set the batch size to 16 and focus on the learning rate, the number of extra attention layers and the weighting coefficient of syntactic loss (for MTS-BioBERT only). Complete hyperparameter grids are summarized in Table 4. Note that ‘-’ indicates that the corresponding parameter is not taken into consideration. We always test parameter combinations in the Cartesian product of the available parameter set: for example, for the Late-Fusion model, we construct 16 ensembles each with one parameter combination out of 16.
Hyperparameter optimization with grid search: customized grid of each model
| Model . | Learning rate . | # extra attention layers . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| SciBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| BioBERT (+extra layers) | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CE-BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CT-BioBERT | |$\left\{ {5{e^{ - 6}},8{e^{ - 6}},1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| Late-Fusion | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| MTS-BioBERT | |$\left\{ {1{e^{ - 5}},5{e^{ - 5}},1{e^{ - 4}}} \right\}$| | – | |$\left\{ {0.1,1.0} \right\}$| |
| Model . | Learning rate . | # extra attention layers . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| SciBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| BioBERT (+extra layers) | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CE-BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CT-BioBERT | |$\left\{ {5{e^{ - 6}},8{e^{ - 6}},1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| Late-Fusion | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| MTS-BioBERT | |$\left\{ {1{e^{ - 5}},5{e^{ - 5}},1{e^{ - 4}}} \right\}$| | – | |$\left\{ {0.1,1.0} \right\}$| |
Hyperparameter optimization with grid search: customized grid of each model
| Model . | Learning rate . | # extra attention layers . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| SciBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| BioBERT (+extra layers) | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CE-BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CT-BioBERT | |$\left\{ {5{e^{ - 6}},8{e^{ - 6}},1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| Late-Fusion | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| MTS-BioBERT | |$\left\{ {1{e^{ - 5}},5{e^{ - 5}},1{e^{ - 4}}} \right\}$| | – | |$\left\{ {0.1,1.0} \right\}$| |
| Model . | Learning rate . | # extra attention layers . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| SciBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| BioBERT (+extra layers) | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CE-BioBERT | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| CT-BioBERT | |$\left\{ {5{e^{ - 6}},8{e^{ - 6}},1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | – | – |
| Late-Fusion | |$\left\{ {1{e^{ - 5}},2{e^{ - 5}},3{e^{ - 5}},5{e^{ - 5}}} \right\}$| | |$\left\{ {1,2,3,4} \right\}$| | – |
| MTS-BioBERT | |$\left\{ {1{e^{ - 5}},5{e^{ - 5}},1{e^{ - 4}}} \right\}$| | – | |$\left\{ {0.1,1.0} \right\}$| |
Evaluation metrics
We use the same evaluation setting as used by the DrugProt organizers (16): we measure micro-averaged precision, recall and F1-score.
Results
In this section, we present the results that we obtain for each of the seven model types: BioBERT, SciBERT, BioBERT (+extra layers), CE-BioBERT, CT-BioBERT, Late-Fusion and MTS-BioBERT. For each model type, we firstly find the optimal combination of hyperparameters based on the average of the micro-averaged F1-scores on the DrugProt validation set. The summary of optimal hyperparameters is shown in Table 5.
Optimal combination of hyperparameters for each model based on the performance on the validation set: learning rate LR, # extra attention layers EAL, |$\alpha $|
| Model . | LR . | EAL . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$2{e^{ - 5}}$| | – | – |
| SciBERT | |$1{e^{ - 5}}$| | – | – |
| BioBERT (+ extra layers) | |$2{e^{ - 5}}$| | 1 | – |
| CE-BioBERT | |$2{e^{ - 5}}$| | 2 | – |
| CT-BioBERT | |$2{e^{ - 5}}$| | – | – |
| Late-Fusion | |$1{e^{ - 5}}$| | 4 | – |
| MTS-BioBERT | |$1{e^{ - 5}}$| | – | 0.1 |
| Model . | LR . | EAL . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$2{e^{ - 5}}$| | – | – |
| SciBERT | |$1{e^{ - 5}}$| | – | – |
| BioBERT (+ extra layers) | |$2{e^{ - 5}}$| | 1 | – |
| CE-BioBERT | |$2{e^{ - 5}}$| | 2 | – |
| CT-BioBERT | |$2{e^{ - 5}}$| | – | – |
| Late-Fusion | |$1{e^{ - 5}}$| | 4 | – |
| MTS-BioBERT | |$1{e^{ - 5}}$| | – | 0.1 |
Optimal combination of hyperparameters for each model based on the performance on the validation set: learning rate LR, # extra attention layers EAL, |$\alpha $|
| Model . | LR . | EAL . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$2{e^{ - 5}}$| | – | – |
| SciBERT | |$1{e^{ - 5}}$| | – | – |
| BioBERT (+ extra layers) | |$2{e^{ - 5}}$| | 1 | – |
| CE-BioBERT | |$2{e^{ - 5}}$| | 2 | – |
| CT-BioBERT | |$2{e^{ - 5}}$| | – | – |
| Late-Fusion | |$1{e^{ - 5}}$| | 4 | – |
| MTS-BioBERT | |$1{e^{ - 5}}$| | – | 0.1 |
| Model . | LR . | EAL . | |$\alpha $| . |
|---|---|---|---|
| BioBERT | |$2{e^{ - 5}}$| | – | – |
| SciBERT | |$1{e^{ - 5}}$| | – | – |
| BioBERT (+ extra layers) | |$2{e^{ - 5}}$| | 1 | – |
| CE-BioBERT | |$2{e^{ - 5}}$| | 2 | – |
| CT-BioBERT | |$2{e^{ - 5}}$| | – | – |
| Late-Fusion | |$1{e^{ - 5}}$| | 4 | – |
| MTS-BioBERT | |$1{e^{ - 5}}$| | – | 0.1 |
We then test each model type on the DrugProt test set using the optimal hyperparameters. We calculate the micro-averaged F1-score for each model within the ensemble and for the voting ensemble. We also compute performance for voting ensembles consisting of different types of model. We merge different types of non-syntax-enhanced models (BioBERT and SciBERT models), as well as both syntax-enhanced and non-syntax-enhanced models. A detailed summary of performances is presented in Table 6. We present voting F1-scores for all models and model combinations; to give an overview of the variance between models in the same ensemble, we also show averaged F1-scores for each model type.
Voting F1-score for each model type (in the second column). %|$\Delta$| denotes the relative gain in voting F1 over BioBERT with no syntax. Averaged F1-score and standard deviation calculated over five independent runs within ensembles for each model type are presented in the rightmost column
| Model . | F1 (vote) . | %|$\Delta$| . | F1 (ave |$ \pm {\rm{ std}}$|) . |
|---|---|---|---|
| Baseline models | |||
| BioBERT | 76.9 | – | 74.3 |$ \pm $| 0.6 |
| SciBERT | 77.0 | +0.1 | 74.0 |$ \pm $| 0.2 |
| BioBERT (+ extra layers) | 76.5 | −0.4 | 74.9 |$ \pm $| 0.2 |
| Syntax-enhanced models | |||
| CE-BioBERT | 75.8 | −1.1 | 74.2 |$ \pm $| 0.6 |
| CT-BioBERT | 75.1 | −1.8 | 72.4 |$ \pm $| 0.5 |
| Late-Fusion | 63.3 | −13.6 | 60.4 |$ \pm $| 1.0 |
| MTS-BioBERT | 70.3 | −6.6 | 66.2 |$ \pm $| 2.2 |
| Combination of non-syntax-enhanced models | |||
| BioBERT + SciBERT | 77.5 | +0.6 | |
| BioBERT + BioBERT (+ extra layers) | 76.7 | −0.2 | |
| Syntax-enhanced + non-syntax-enhanced models | |||
| BioBERT + CE-BioBERT | 76.5 | −0.4 | |
| BioBERT + CT-BioBERT | 76.7 | −0.2 | |
| BioBERT + Late-Fusion | 72.9 | −4.0 | |
| BioBERT + MTS-BioBERT | 74.9 | −2.0 | |
| Model . | F1 (vote) . | %|$\Delta$| . | F1 (ave |$ \pm {\rm{ std}}$|) . |
|---|---|---|---|
| Baseline models | |||
| BioBERT | 76.9 | – | 74.3 |$ \pm $| 0.6 |
| SciBERT | 77.0 | +0.1 | 74.0 |$ \pm $| 0.2 |
| BioBERT (+ extra layers) | 76.5 | −0.4 | 74.9 |$ \pm $| 0.2 |
| Syntax-enhanced models | |||
| CE-BioBERT | 75.8 | −1.1 | 74.2 |$ \pm $| 0.6 |
| CT-BioBERT | 75.1 | −1.8 | 72.4 |$ \pm $| 0.5 |
| Late-Fusion | 63.3 | −13.6 | 60.4 |$ \pm $| 1.0 |
| MTS-BioBERT | 70.3 | −6.6 | 66.2 |$ \pm $| 2.2 |
| Combination of non-syntax-enhanced models | |||
| BioBERT + SciBERT | 77.5 | +0.6 | |
| BioBERT + BioBERT (+ extra layers) | 76.7 | −0.2 | |
| Syntax-enhanced + non-syntax-enhanced models | |||
| BioBERT + CE-BioBERT | 76.5 | −0.4 | |
| BioBERT + CT-BioBERT | 76.7 | −0.2 | |
| BioBERT + Late-Fusion | 72.9 | −4.0 | |
| BioBERT + MTS-BioBERT | 74.9 | −2.0 | |
Voting F1-score for each model type (in the second column). %|$\Delta$| denotes the relative gain in voting F1 over BioBERT with no syntax. Averaged F1-score and standard deviation calculated over five independent runs within ensembles for each model type are presented in the rightmost column
| Model . | F1 (vote) . | %|$\Delta$| . | F1 (ave |$ \pm {\rm{ std}}$|) . |
|---|---|---|---|
| Baseline models | |||
| BioBERT | 76.9 | – | 74.3 |$ \pm $| 0.6 |
| SciBERT | 77.0 | +0.1 | 74.0 |$ \pm $| 0.2 |
| BioBERT (+ extra layers) | 76.5 | −0.4 | 74.9 |$ \pm $| 0.2 |
| Syntax-enhanced models | |||
| CE-BioBERT | 75.8 | −1.1 | 74.2 |$ \pm $| 0.6 |
| CT-BioBERT | 75.1 | −1.8 | 72.4 |$ \pm $| 0.5 |
| Late-Fusion | 63.3 | −13.6 | 60.4 |$ \pm $| 1.0 |
| MTS-BioBERT | 70.3 | −6.6 | 66.2 |$ \pm $| 2.2 |
| Combination of non-syntax-enhanced models | |||
| BioBERT + SciBERT | 77.5 | +0.6 | |
| BioBERT + BioBERT (+ extra layers) | 76.7 | −0.2 | |
| Syntax-enhanced + non-syntax-enhanced models | |||
| BioBERT + CE-BioBERT | 76.5 | −0.4 | |
| BioBERT + CT-BioBERT | 76.7 | −0.2 | |
| BioBERT + Late-Fusion | 72.9 | −4.0 | |
| BioBERT + MTS-BioBERT | 74.9 | −2.0 | |
| Model . | F1 (vote) . | %|$\Delta$| . | F1 (ave |$ \pm {\rm{ std}}$|) . |
|---|---|---|---|
| Baseline models | |||
| BioBERT | 76.9 | – | 74.3 |$ \pm $| 0.6 |
| SciBERT | 77.0 | +0.1 | 74.0 |$ \pm $| 0.2 |
| BioBERT (+ extra layers) | 76.5 | −0.4 | 74.9 |$ \pm $| 0.2 |
| Syntax-enhanced models | |||
| CE-BioBERT | 75.8 | −1.1 | 74.2 |$ \pm $| 0.6 |
| CT-BioBERT | 75.1 | −1.8 | 72.4 |$ \pm $| 0.5 |
| Late-Fusion | 63.3 | −13.6 | 60.4 |$ \pm $| 1.0 |
| MTS-BioBERT | 70.3 | −6.6 | 66.2 |$ \pm $| 2.2 |
| Combination of non-syntax-enhanced models | |||
| BioBERT + SciBERT | 77.5 | +0.6 | |
| BioBERT + BioBERT (+ extra layers) | 76.7 | −0.2 | |
| Syntax-enhanced + non-syntax-enhanced models | |||
| BioBERT + CE-BioBERT | 76.5 | −0.4 | |
| BioBERT + CT-BioBERT | 76.7 | −0.2 | |
| BioBERT + Late-Fusion | 72.9 | −4.0 | |
| BioBERT + MTS-BioBERT | 74.9 | −2.0 | |
Baseline BioBERT and SciBERT models both obtain high F1-scores (respectively, 76.9 and 77.0). Among the syntax-enhanced models, CE-BioBERT gives the best performance (75.8), although it is unable to outperform baseline models. Combining BioBERT and SciBERT baseline models improves over individual models and yields the highest performance overall (77.5). Combining syntax-enhanced and non-syntax-enhanced also improves over individual syntax-enhanced models but performance remains below that of non-syntax-enhanced models.
Discussion
We can see from the results in the previous section that in all the tested syntax-enhanced models, introducing syntax degrades, to varying degrees, the performance of biomedical RE summarized in Table 6. Besides, integrating dependency information has a more significant negative impact than integrating constituency information. Quite different from what is observed by Sachan et al. (7) in their experiments (they use Stanford CoreNLP as syntactic parser), our results show that replacing the original word-to-word attention matrix by a hard-pruned adjacency matrix of the dependency graph causes severe degradation on DrugProt, rather than slightly boosting the performance.
Impact of syntactic parsing quality
We make a hypothesis that the degradation of syntax-enhanced models might be due in part to the quality of dependency information obtained from the off-the-shelf syntactic parser Stanza on the DrugProt corpus.
To verify this hypothesis, we manually evaluated dependency analysis on a small subset of sentences in the validation set. Firstly, we divide sentences in the validation set into groups according to the length of the shortest dependency path (SDP) between the two arguments of the candidate relation. We then randomly select sentences from each group to form a subset containing 48 sentences. For each sentence, we manually examine the dependency parse provided by Stanza and report two numbers: the number of erroneous dependency links on the full-dependency parses and the number of erroneous dependency links in the SDP between the two arguments of the candidate relation. The number of errors on the full-dependency parses allowed us to compute an ‘Unlabelled Attachment’ Precision score of 94.9. Compared to Stanza (17) reporting 91.01 as Unlabeled Attachment Score on the GENIA treebank (18), although we cannot make a direct comparison (since their score is a F1-score), it is fair to conclude that the quality of dependency parses from Stanza is acceptable on the subset of sentences that we extracted.
We further analyse whether there exists a negative correlation between the number of errors (in full sentences and in SDPs) in the dependency parsing and the RE performance of Late-Fusion and MTS-BioBERT, which are enhanced with dependency information. We gather sentences that share the same number of errors and calculate micro F1-scores on sentences from each group. Note that ‘no_relation’ is not counted in the calculation of micro F1-scores, so we remove groups that include only examples labelled as ‘no_relation’ (otherwise, in the stratified results zero micro F1-scores will be reported for these groups which are unrepresentative). Because only Late-Fusion and MTS-BioBERT integrate dependency information, we analyse only these two models. This result is visualized in Figure 6. To highlight the significance of results for each group, the width of the bars is proportional to the number of examples for each group. We observe that both figures show no negative correlation.
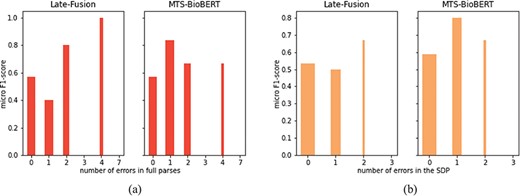
RE performance on the subset of sentences used for manual dependency analysis: (a) stratified by the number of erroneous dependency links identified in full parses and (b) stratified by the number of erroneous dependency links in SDPs.
Mathematically, the size of the subset on which we perform manual analysis is relatively small and there exist outliers: for example, Late-Fusion obtains 1.0 F1-score on sentences with four total errors, and obtains 0.0 F1-score with three errors in the SDP. So we choose to compute Spearman’s rank coefficient between the numbers of errors and corresponding micro F1-scores. The result is summarized in Table 7. Spearman’s rank coefficients indicate that there is no strong negative correlation in either cases (high P-values indicate that two variables are uncorrelated, which is the null hypothesis in the computation of Spearman’s rank coefficient), which is coherent with our previous observation from the figures. Therefore, we conclude that errors in the dependency parse do not lead to degradation of RE performance. This may be explained by the fact that errors in dependency parses are often ‘local’, and our proposed syntax-enhanced models search to capture the linkage of full-dependency graph or geometric properties of full-dependency trees. To summarize, our hypothesis about relating degradation of syntax-enhanced models to the quality of syntactic parsing does not hold, since by manually examining dependency parses of Stanza on a randomly selected subset of sentences, we observe a rather good decision; at the same time, there is no strong negative correlation between the number of errors in the dependency parsing (either in full sentences or in SDPs) and the RE performance.
Spearman’s rank coefficient |${r_s}$| between number of errors (in full sentences and in SDPs) and micro F1-score that syntax-enhanced models obtain
| Model . | rs . | P-value . |
|---|---|---|
| # total errors | ||
| Late-Fusion | −0.20 | 0.70 |
| MTS-BioBERT | −0.44 | 0.38 |
| # errors in the SDP | ||
| Late-Fusion | −0.40 | 0.60 |
| MTS-BioBERT | −0.40 | 0.60 |
| Model . | rs . | P-value . |
|---|---|---|
| # total errors | ||
| Late-Fusion | −0.20 | 0.70 |
| MTS-BioBERT | −0.44 | 0.38 |
| # errors in the SDP | ||
| Late-Fusion | −0.40 | 0.60 |
| MTS-BioBERT | −0.40 | 0.60 |
Spearman’s rank coefficient |${r_s}$| between number of errors (in full sentences and in SDPs) and micro F1-score that syntax-enhanced models obtain
| Model . | rs . | P-value . |
|---|---|---|
| # total errors | ||
| Late-Fusion | −0.20 | 0.70 |
| MTS-BioBERT | −0.44 | 0.38 |
| # errors in the SDP | ||
| Late-Fusion | −0.40 | 0.60 |
| MTS-BioBERT | −0.40 | 0.60 |
| Model . | rs . | P-value . |
|---|---|---|
| # total errors | ||
| Late-Fusion | −0.20 | 0.70 |
| MTS-BioBERT | −0.44 | 0.38 |
| # errors in the SDP | ||
| Late-Fusion | −0.40 | 0.60 |
| MTS-BioBERT | −0.40 | 0.60 |
However, due to time limitations, we have focused our analysis on dependency parses. To be more thorough and give a more definitive conclusion on the impact of parsing quality, analysis on constituency parses should also be performed. We plan to do this in future work.
Training difficulties
Besides the quality of syntactic parsers, we also hypothesize that the degradation in performance of the CT-BioBERT and MTS-BioBERT models may be due to the difficulty of training neural networks. We had speculated that constituency tag information and hierarchical information of the constituency tree encoded in the linearized sequence would compensate the possible degradation caused by adding constituent tags as new wordpieces, but this hypothesis is not supported by the results. For MTS-BioBERT, using a same learning rate to jointly train the neural network on three tasks may not be ideal to make a balance between three training objectives.
Difference between syntax-enhanced models and BioBERT
Although we observe degraded performance of syntax-enhanced models, it remains unclear how these models behave differently compared to BioBERT. We examine the following questions: although syntax degrades performance in general, (i) do syntax-enhanced models make unique errors that BioBERT does not make and vice versa? and (ii) do syntax-enhanced models improve performance on some examples?
To answer the first question, we choose the best syntax-enhanced model CE-BioBERT and find mistakes in the validation set that occur solely for CE-BioBERT and BioBERT, respectively. There exist 212 CE-BioBERT-only mistakes and 272 BioBERT-only mistakes. Because it is hard to manually examine these examples and find the differences, we choose to calculate statistics for the two groups of mistakes: surface distance between the arguments of a candidate relation; the length of the dependency path between the subject and the object of a candidate relation and sentence length. The result is shown in Table 8. We observe that BioBERT make mistakes on examples which consist of slightly longer sentences and longer distance between the arguments. This observation leads us to hypothesize that CE-BioBERT might perform better than BioBERT in cases where inter-argument distance of the target relation is longer.
Statistics on mistakes that only occur for CE-BioBERT and only for BioBERT. Surface distance refers to inter-argument distance, and SDP length refers to the length of the SDP between arguments
| Model . | Surface distance . | SDP length . | Sentence length . |
|---|---|---|---|
| BioBERT | 17.2 | 4.8 | 43.7 |
| CE-BioBERT | 16.3 | 4.5 | 43.0 |
| Model . | Surface distance . | SDP length . | Sentence length . |
|---|---|---|---|
| BioBERT | 17.2 | 4.8 | 43.7 |
| CE-BioBERT | 16.3 | 4.5 | 43.0 |
Statistics on mistakes that only occur for CE-BioBERT and only for BioBERT. Surface distance refers to inter-argument distance, and SDP length refers to the length of the SDP between arguments
| Model . | Surface distance . | SDP length . | Sentence length . |
|---|---|---|---|
| BioBERT | 17.2 | 4.8 | 43.7 |
| CE-BioBERT | 16.3 | 4.5 | 43.0 |
| Model . | Surface distance . | SDP length . | Sentence length . |
|---|---|---|---|
| BioBERT | 17.2 | 4.8 | 43.7 |
| CE-BioBERT | 16.3 | 4.5 | 43.0 |
To answer the second question, from our previous observation we hypothesize that syntax may help identify relations on examples in which two arguments of a candidate relation are far from each other, because the subject–object distance of the target relation is shortened due to integrated dependency information (by a shortened syntactic path) or constituency information (by grouping words into constituents). We thus stratify examples in the validation set based on subject–object surface distance (e.g. if the subject is before the object, the subject–object surface distance equals the number of tokens between the last token of the subject and the first token of the object) and then compare the performance of BioBERT and of all syntax-enhanced models. We first divide all possible distances into intervals to get a general overview of the stratified results. The result is shown in Figure 7. It is worth noting that for examples in which the subject and the object refer to the same token, there are 1153/1167 examples labelled ‘no_relation’ (true negatives), and this label is excluded in the calculation of the micro F1-score. Besides, we observe that CE-BioBERT performs better than BioBERT in multiple cases, which is coherent with the fact that CE-BioBERT outperforms BioBERT on the validation set (which is not the case on the test set). We observe also that when the subject–object surface distance is superior or equal to 40, CE-BioBERT, CT-BioBERT and Late-Fusion outperform BioBERT, which validates our hypothesis about possible improvements brought by syntax-enhanced models in cases where the distance between two arguments of candidate relation is long. We further subdivide the interval ‘superior or equal to 40’ into five intervals and show the result in Figure 8. We observe that for each interval, syntax-enhanced models outperform BioBERT, which confirms that syntax helps improve the performance of RE on examples in which the subject–object surface distance is relatively long (|$ \ge $|40). However, due to the limited number of these examples (|$ \approx $|2.7%), the overall performance of syntax-enhanced models is worse than BioBERT. This gives us a clue towards building a hybrid system consisting of BioBERT and syntax-enhanced models and uses them respectively on examples with short-distance arguments and long-distance examples during the inference stage. Because labels of the DrugProt test set are unavailable and there exists a huge amount of dummy data in the test set, it is impossible to know how many long-distance examples are used for evaluation, and we leave this part for future work.
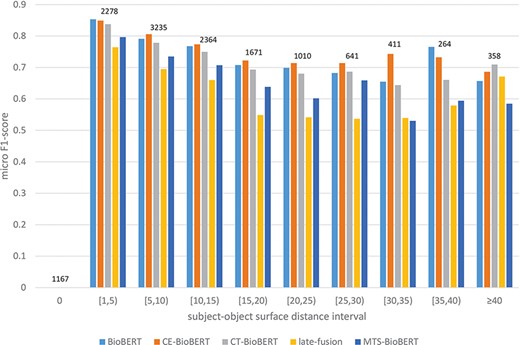
Stratified results on DrugProt validation set: Examples in the validation set are regrouped based on their subject–object surface distances. Intervals are of length 5 except two special cases 0 and |$ \ge $|40. For each interval, the number of examples that fall in this interval is shown on top of bars.
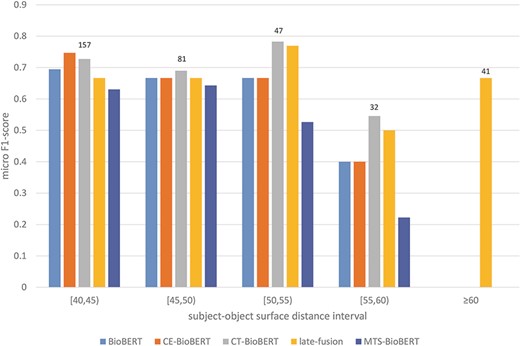
Stratified results on the DrugProt validation set (surface distance |$ \ge $|40): Examples in the validation set are regrouped based on their subject–object surface distances. For each interval, the number of examples that fall in this interval is shown on top of the bars.
Conclusion
In this work, we investigate the effect of integrating syntactic information into pre-trained BioBERT models on a chemical–drug RE task. We conduct experiments with our proposed syntax-enhanced architectures CE-BioBERT, CT-BioBERT and MTS-BioBERT, along with an existing syntax-enhanced architecture (Late-Fusion) on the DrugProt corpus from the BioCreative VII track 1 shared task. By comparing the performance of syntax-enhanced architectures with three baseline models, we find that these syntax-enhanced models degrade, to varying degrees, the performance of BioBERT in general. We also investigate a hypothesis relating the quality of syntactic parses and the degradation of RE performance by manually analysing dependencies on a randomly selected sentence from the DrugProt validation set. The result shows that there is no negative correlation between the number of errors in dependency parses obtained from an off-the-shelf parser Stanza and the RE performance. Although syntax-enhanced models are found to degrade the general RE performance, stratified results on the validation set show that syntax-enhanced models improve the performance of RE on examples in which the subject–object (arguments of the target relation) surface distance is long (|$ \ge $|40). In future work, we will investigate more thoroughly the impact of parsing quality, suggest to build hybrid systems that ensemble both BioBERT and syntax-enhanced models and choose different models at inference stage based on subject–object surface distances. We also plan to test our models and perform analyses on other specialized corpora besides the DrugProt corpus. This will allow us to draw more nuanced conclusions regarding the injection of syntax into BERT models for RE in the biomedical domain. Additionally, we plan to look at other types of information to integrate into the models, such as information from biomedical knowledge bases, which might be more stable and more reliable than syntactic parsing.
Acknowledgements
We are grateful to the Saclay-IA platform of Université Paris-Saclay for providing computing and storage resources through its Lab-IA GPU cluster.
Funding
Labex DigiCosme [project ANR-11-LABEX-0045-DIGICOSME] operated by ANR as part of the program ‘Investissement d’Avenir’ Idex Paris-Saclay [ANR-11-IDEX-0003-02].
Conflict of interest
There is no competing interest.



