-
PDF
- Split View
-
Views
-
Cite
Cite
Wanying Dou, Hemn Barzan Abdalla, Xu Chen, Changyi Sun, Xuefei Chen, Qiwen Tian, Junyi Wang, Wei Zhou, Wei Chi, Xuan Zhou, Hailv Ye, Chuyun Bi, Xuechen Tian, Yixin Yang, Aloysius Wong, ProbResist: a database for drug-resistant probiotic bacteria, Database, Volume 2022, 2022, baac064, https://doi.org/10.1093/database/baac064
Close - Share Icon Share
Abstract
Drug resistance remains a global threat, and the rising trend of consuming probiotic-containing foods, many of which harbor antibiotic resistant determinants, has raised serious health concerns. Currently, the lack of accessibility to location-, drug- and species-specific information of drug-resistant probiotics has hampered efforts to combat the global spread of drug resistance. Here, we describe the development of ProbResist, which is a manually curated online database that catalogs reports of probiotic bacteria that have been experimentally proven to be resistant to antibiotics. ProbResist allows users to search for information of drug resistance in probiotics by querying with the names of the bacteria, antibiotic or location. Retrieved results are presented in a downloadable table format containing the names of the antibiotic, probiotic species, resistant determinants, region where the study was conducted and digital article identifiers (PubMed Identifier and Digital Object Identifier) hyperlinked to the original sources. The webserver also presents a simple analysis of information stored in the database. Given the increasing reports of drug-resistant probiotics, an exclusive database is necessary to catalog them in one platform. It will enable medical practitioners and experts involved in policy making to access this information quickly and conveniently, thus contributing toward the broader goal of combating drug resistance.
Introduction
Probiotics are live microorganisms that confer a broad range of health benefits to the host if consumed in adequate amounts (1). The health-promoting properties include modulating the immune system and treating inflammatory diseases (2, 3), preventing cancer and acting as adjuvant for chemotherapy (4), treating viral respiratory infections (5, 6), reducing low-density lipoprotein cholesterol and improving risk factors associated with coronary heart disease (7), eliciting beneficial metabolic effects in patients with type 2 diabetes mellitus and improving blood lipid profiles (8), assisting the treatment and management of oral diseases including halitosis, dental caries and periodontitis (9, 10) as well as regulating anxiety, mood, cognition and pain, and treating and preventing neurologic disorders (11–13). As the health claims of probiotics continue to emerge, their number of applications also increases, as exemplified by the recent growth in the global probiotic market and the development of new probiotic-containing foods, supplements and health products (14–20). However, it is noted that in Europe, the European Food Safety Agency does not permit the term ‘probiotic’ to be used in foods and food supplements (21).
There are overwhelming reports of probiotic bacteria isolated from various foods, animal and human samples that are resistant to antibiotics used in clinical and veterinary applications such as vancomycin, metronidazole, chloramphenicol, erythromycin, quinupristin/dalfopristin, lincomycin, clindamycin and tetracyclines (16, 17, 22–25). Moreover, the resistant determinants responsible for the resistant phenotypes such as blaZ and mecA for beta-lactam antibiotics; aac(6′)-aph(2′′), ant(6) and aph(3′)-IIIa for aminoglycosides; erm(A), erm(B), erm(C), mefA and lnuA for erythromycin and tet(W), tet(L), tet(K), tet(S) and tet(M) for tetracycline have been characterized (26–33). While drug-resistant probiotics may not cause direct harm to humans, they can however transfer their resistant determinants not only to commensals but also to pathogens in the human gut or the oral cavity. Multiple studies have raised the concern that long-term consumption of foods containing drug-resistant probiotics will establish a reservoir of drug-resistant determinants in the gut or oral cavity, and risk acquisitions through horizontal gene transfer by opportunistic pathogens (16, 17, 33–45). If so, it will render antibiotic treatments ineffective, thus exacerbating the problem of drug resistance in the clinical settings such as when recommending treatments for life-threatening diseases caused by pathogens, when treating immunocompromised patients or when performing surgical procedures (46–50).
There is increasing evidence supporting the health risks of excessive or long-term consumption of drug-resistant probiotics. For instance, experimental evidence has demonstrated the conjugal transfer of resistant genes such as the erythromycin-resistant plasmid pLFE1 and tetracycline-resistance gene tet(M) from one probiotic strain to another of the same genus and crucially also to pathogenic strains such as Listeria innocua, Listeria monocytogenes and Enterococcus faecalis (51, 52). In addition to in vitro filter mating studies, conjugal transfer of resistant genes from probiotics to pathogens in vivo, in the animal gut, and in fermentation stages during food processing, have also been reported (32). Recent evidence from metagenomics studies linking probiotic consumption to the increase in the number of resistant genes as well as the number of strains carrying the resistant genes has also been reported in mice and humans (53). Meanwhile, population genetic studies from selected Asian and European cohorts have also identified antibiotic-resistant genes in the human gut that are highly identical to known resistant genes of antibiotics widely used in animal husbandry (54). Notably, antibiotics from food and environmental sources were thought to have greater impact in establishing a reservoir of resistant genes in the human gut than antibiotics used in human medicine (55). Considering this evidence and given the projection that by 2050, more people will succumb to drug resistance-related illnesses than cancer (56), the health risk of drug-resistant probiotics has become even more concerning.
Currently, the lack of accessibility to location-, drug- and organism-specific information of drug resistance in probiotic bacteria has hampered efforts to combat the global spread of drug resistance. Here, we built an online-based database that allows users to search for raw and processed information of drug resistance in probiotics quickly and conveniently, by querying with the names of the bacteria, antibiotic or location. A comprehensive catalog of drug-resistant probiotic bacteria can serve medical practitioners in providing better health care services including treatment, prescriptions and/or prevention advice to patients and inform experts involved in policy making, thus the development of ProbResist.
Materials and methods
Selection of bacteria for inclusion in ProbResist database
Relevant search terms such as ‘antibiotic resistance’, ‘drug resistance’, ‘probiotics’ and the combination thereof were used to search for articles reporting on drug-resistant probiotics from the PubMed database. In addition to the previous Lactobacillus genus name, the new names ‘Holzapfelia’, ‘Amylolactobacillus’, ‘Bombilactobacillus’, ‘Companilactobacillus’, ‘Lapidilactobacillus’, ‘Agrilactobacillus’, ‘Schleiferilactobacillus’, ‘Loigolactobacilus’, ‘Lacticaseibacillus’, ‘Latilactobacillus’, ‘Dellaglioa’, ‘Liquorilactobacillus’, ‘Ligilactobacillus’, ‘Lactiplantibacillus’, ‘Furfurilactobacillus’, ‘Paucilactobacillus’, ‘Limosilactobacillus’, ‘Fructilactobacillus’, ‘Acetilactobacillus’, ‘Apilactobacillus’, ‘Levilactobacillus’, ‘Secundilactobacillus’, ‘Lentilactobacillus’, ‘Lactobacillaceae’ and ‘Leuconostocaceae’ were also used as keywords together with the terms ‘antibiotic resistance’ or ‘drug resistance’, in our search (57) (Supplementary Table S1). The retrieved articles were manually curated where, e.g. review articles were excluded to avoid overlapping reports of original research, before extracting information of probiotic strains reported, geographical location of study, antibiotics which were tested and the resistant genes, for inclusion in ProbResist database.
Information included in ProbResist database
ProbResist database contains the drug-resistant probiotic strains, the geographical location where drug resistance was reported, the antibiotics which were tested, the source of the probiotic isolates and the nature of the resistant genes, if known. The article electronic identification numbers [PubMed Identifier (PMID) and Digital Object Identifier (DOI)] were also included in the database and hyperlinked to their original sources. An abbreviation legend of the bacteria names, e.g. Bb. = Bifidobacterium, Sc. = Streptococcus and W. = Weissela, was provided for easy referencing and clarity. The new names of Lactobacillus such as Lacticaseibacillus casei or Limosilactobacillus reuteri are spelled out in full. The database is periodically curated and updated to include new reports of drug-resistant probiotics.
ProbResist database construction and interface
Search terms such as ‘antibiotic resistance’, ‘drug resistance’ and ‘probiotics’ and their combinations (see keywords under the section: selection of bacteria for inclusion in ProbResist database) were used to search for relevant reports from PubMed database, and the information, e.g. probiotic strains, location, source of the isolates, antibiotics, and nature of the resistant genes, was extracted to build the database for ProbResist. The extracted information is primally curated and formatted in Excel. The formatted sheet is then imported into MySQL database in the following format: antibiotic, species, gene location, source of isolates, the region or country where the study was conducted, and article electronic identification numbers (PMID and DOI). The front-end and back-end separation of the database website are programmed through a JavaScript Object Notation (JSON) format interface. The front-end is responsible for calling Asynchronous JavaScript and XML (AJAX) to fetch data and display, and the back-end provides Application Programming Interface (API) interface, and corresponding calculations. The back-end provides an interface to the front-end, and the front-end displays data according to the interface. When the user searches with the names of the probiotic species, antibiotic, or country, the front-end calls the database while the back-end provides the data in JSON format (Figure 1). The web template from HTML design (https://html.design) is adopted and modulated.
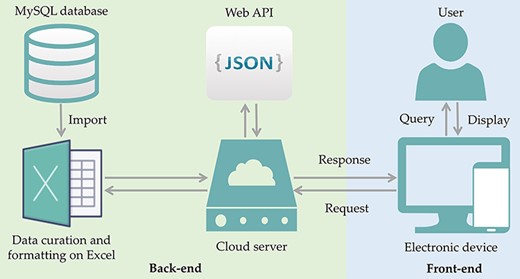
ProbResist database and interface. Information extracted from articles retrieved from PubMed database was manually curated and formatted offline on Excel before importing into MySQL database in the following format: antibiotic, species, gene location, country, PMID and DOI. The database website structure was designed and developed using the front-end (blue box) and back-end (green box) separation where the website front-end will query the database when user invoked either the search function by species, antibiotic or country name, in the search bar. At the back-end, data are provided by restful web API in JSON format. For website development, an online web template provided by HTML design (https://html.design) was adopted and modulated. Front-end only requires the setting up of the user query input as a parameter to specify API URL and then displayed in appropriate table HTML format. Users have the option to export their retrieved results into Excel format.
Results
ProbResist is available at https://probresist.com without registration or license. This database catalogs published reports of probiotic bacteria from various sources that have been experimentally proven to be resistant to one or more antibiotics. As ProbResist gathers relevant information in one website, it provides quick and convenient access to processed information on drug resistance in probiotics. In addition to the browse functions, the webserver also provides simple analysis of the database in graphical formats. Specifically, it presents (i) the breakdown of the type of probiotic bacteria most frequently reported to be resistant to antibiotics as pie chart, (ii) the abundance of antibiotic resistance reported in the scientific literature for probiotic bacteria as bar graph, (iii) the antibiotic classes, generations and mode of actions as tables and (iv) the frequency of reports of probiotics from different regions of countries that are resistant to antibiotics as heat map.
The website features a top aligned white banner that houses the name of the database on the left and a browse full database tab on the right. Following the banner is a text search box accompanied by brief instructions that allow experienced users to quickly query and retrieve data from ProbResist. The Lactobacillus genus has been recently reclassified into more than 25 genera including 23 novel genera, thus newer articles (after April 2020) normally adopt these new names in their reports (57). To retrieve both the older and newer articles in the ProbResist database, specific instructions are provided on the website beneath the main search bar. Users can use the abbreviated forms of the bacteria, e.g. B. = Bacillus; Lb. = Lactobacillus when searching for older articles containing the previous Lactobacillus names but for newer articles, users are required to use the full names, e.g. Lacticaseibacillus casei or Limosilactobacillus reuteri. For convenience, a list of prominent Lactobacillus probiotic species and their new names, e.g. Lb. casei = Lacticaseibacillus casei; Lb. paracasei = Lacticaseibacillus paracasei; Lb. rhamnosus = Lacticaseibacillus rhamnosus; Lb. plantarum = Lactiplantibacillus plantarum; Lb. brevis = Levilactobacillus brevis; Lb. salivarius = Ligilactobacillus salivarius; Lb. fermentum = Limosilactobacillus fermentum and Lb. reuteri = Limosilactobacillus reuteri, is also provided. Direct links to the announcement by the International Scientific Association for Probiotics and Prebiotics (https://isappscience.org/new-names-for-important-probiotic-lactobacillus-species/) and the published article (57) are also provided for those who wish to learn more about the Lactobacillus taxonomy changes. The search box allows users to query the database from three search options: antibiotic, species or region/country name. Since the database stores the abbreviated form of the bacteria, a legend of the abbreviated bacteria is provided beneath the search box for easy referencing (Figure 2). To search for newer articles containing the new Lactobacillus names, their full forms must be used. The search box implements the autocomplete function with a drop-down menu containing text suggestions to aid users identify their search terms.
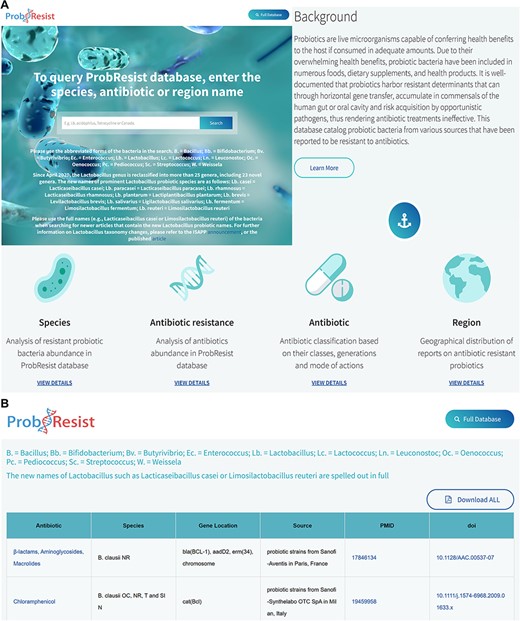
Layout of the ProbResist webserver. (A) Top panel: screenshot of the home page beginning with a text search bar that allows experienced users to quickly query the database from three search options: bacteria species, antibiotic or region/country name. The search bar is accompanied by a detailed instruction and species abbreviation legend to aid user input. Following that, an introduction section containing a brief description of probiotics and antibiotic resistance is provided to aid first time users navigate and extract information from the database. Bottom panel: the next part of the webserver contains general information of the antibiotics as well as a brief analysis of the database organized based on the frequency of the respective species, drug resistance, antibiotic classes and region/country of study. (B) In the result page, retrieved results contain the name of antibiotic, bacteria species, gene name and location, source of isolates, country of study and the unique digital article identifiers PMID and DOI, hyperlinked to the original sources. The website allows users to export their results as Excel.
Following the search box is the background information containing a description of probiotics and drug resistance to explain the nature and utility of the database in more detail, which aids first-time users navigate and extract information from the database. A ‘learn more’ tab beneath the background information links directly to selected articles, which provide users with more information from the literature regarding the health impact and recent developments of drug-resistant probiotics. Following the background information is a brief analysis section containing species, antibiotic resistance, antibiotic classes and geographical location, each represented by corresponding logos. This section allows user to assess processed information of ProbResist database in graphical formats such as the frequency of reports according to the bacteria species, drug resistance, antibiotic classes and geographical location. The website allows users to download the dataset of their searches in Excel format (Figure 2).
As of May 2022, ProbResist has 158 articles reporting on drug-resistant probiotics in its database. A brief analysis of the database metrics is provided on the website following the introduction section. In the analysis of resistant probiotic bacteria abundance in ProbResist database, Lactobacillus including their new names (744 times) is by far the largest group of probiotic bacteria reported to be resistant to antibiotics, followed by Bifidobacterium (81 times), Pediococcus (44 times), Weissela (36 times), Lactococcus (23 times), Streptococcus (18 times) and Bacillus (13 times), while the analysis of antibiotic types reported that probiotics resistant to tetracycline (91 times) is detected most frequently in the literature followed by erythromycin (70 times), vancomycin (63 times), streptomycin (60 times), chloramphenicol (47 times), kanamycin (40 times), ampicillin (37 times) and gentamicin (36 times). As for the analysis of the geographical distribution of reports on antibiotic-resistant probiotics, China, with 22 scientific articles, has the highest frequency of reports on antibiotic-resistant probiotics globally, followed by Belgium with 20, Italy with 19 and US and Spain, each with 10 articles, respectively (Supplementary Table S1).
In the result page, retrieved data are presented in a table containing the name of antibiotic, the bacteria species, the name of the resistant determinants and their locations whether on plasmids, transposons or chromosomes, the source of probiotic isolates, the region or country where the study was conducted, as well as the unique article identifiers PMID and DOI, with hyperlinks linking directly to the original sources (Figure 2). The antibiotics, which probiotics are reported to be resistant to, are also hyperlinked to the antibiotic table containing information of the classes, generations and mode of actions. Above the table, a legend containing abbreviations of the bacteria names is provided and users are also reminded that the new names of Lactobacillus such as Lacticaseibacillus casei or Limosilactobacillus reuteri are spelled out in full in the database. At the bottom panel of the webserver is contact information as well as the current development status and planned updates of the webserver. Users have the option to export the retrieved results as Excel by clicking the ‘download all’ tab at the top of the table.
Discussion
Recent authoritative reviews on probiotic safety have also focused on the risk of probiotics in trafficking resistant determinants among other adverse effects such as causing systemic infections in immunocompromised individuals and deleterious metabolic activities (17, 34–37, 43, 58–63). Antibiotics especially those from multiple classes such as penicillins, cephalosporins, sulfonamides and macrolides prescribed to infants have been associated with increased risk for allergy including food allergy, atopic and contact dermatitis, allergic rhinitis and allergic conjunctivitis, anaphylaxis and asthma, during the later stage of development. This is likely achieved through the disruption of the antibiotic-associated microbiome diversity (64, 65). Indeed, it was shown in mice that antibiotic-associated reduction in intestinal microbiota and metabolic abundance such as short-chain fatty acids and tryptophan resulted in an increase in inflammatory response as evidenced by elevated immunoglobulins IgE and IgG1 after sensitization. Upon re-exposure to the allergens, the mice showed evidence for damaged intestinal barrier such as ruptured intestinal villi and a decrease in tight junction proteins (66). Thus, excessive or irresponsible use of probiotics, many of which are phenotypically resistant to clinically important antibiotics and harbor resistance genes on mobile genetic elements, could further exacerbate the current public health crisis especially in the clinical settings where those with underlying health conditions are most vulnerable. For instance, a meta-analysis study across 15 different countries revealed that diabetes mellitus has a significant association with multidrug-resistant tuberculosis (67), while patients with chronic kidney disease are likely to be the reservoir of antibiotic-resistant pathogens as multidrug-resistant organisms such as vancomycin-resistant enterococci or methicillin-resistant Staphylococcus aureus are prevalent in these individuals (68). On the other hand, drug-resistant human immunodeficiency virus (HIV) is already recognized as an emerging threat to epidemic control since HIV patients more frequently contract opportunistic infections including tuberculosis and have higher risk for Coronavirus Disease 2019 (COVID-19) mortality (69–71).
To our knowledge, ProbResist is the first and most comprehensive webtool to catalog probiotic bacteria globally that have been reported to be resistant to antibiotics. Since drug-resistant probiotics are relatively understudied compared to clinically relevant or disease-causing strains, a more focused probiotic-specific database would allow this increasingly popular group of bacteria to be highlighted especially since there is increasing demand corresponding to the rising trend of dietary, health and food supplement use (15, 62–74). The latter contains the highest amount of probiotic bacteria per serving compared to other foods, and recent reports of drug resistance in probiotics from health supplements further exacerbate its health concerns (17, 34, 35). Due to its fungal nature, yeast such as Saccharomyces boulardii is intrinsically resistant to antibiotics and is already shown to exhibit some probiotic properties such as alleviating infections in the gut. Thus, as alternatives to lactic acid bacteria, commensal fungal species could be further explored and developed using modern genetic engineering methods to produce effective and safer probiotics (75, 76).
ProbResist is an initial effort to catalog probiotic bacteria that have been reported in scholarly articles to be resistant to drugs. Since this database only includes reports from published articles cataloged in PubMed, it will require a community-driven effort to make this database as comprehensive as possible as more localized reports and community hospital data especially those in the developing and underdeveloped countries, remain unpublished. As such, users are encouraged to reach us through the email provided at the bottom of the webpage to propose unreported drug-resistant probiotics that warrant inclusion in our database and our team will validate the request and update the database accordingly. Additionally, we are also exploring other repositories to identify more reports of drug-resistant probiotics and conduct periodic updates on the database as new experimental evidence surface.
Conclusion
Given the increasing number of reported drug-resistant probiotics (77, 78), an exclusive database is therefore necessary to organize reports of drug-resistant probiotics in one platform, which enables health professionals, medical practitioners and experts involved in policy making, to quickly and conveniently search for, and assess the processed forms of, this information. The increasing connectivity especially in developing countries also enables our webserver to contribute towards the broader goal of combating antibiotic resistance.
Supplementary data
Supplementary data are available at Database Online.
Author contributions
Conceptualization, A.W.; methodology, A.W., W.D. and H.B.A.; formal analysis, A.W. and W.D., investigation, A.W., W.D., Xu.C., C.S., Xue.C., Q.T. and J.Y.; resources, A.W., X.Z., H.Y., C.B., X.T. and Y.Y.; data curation, A.W., W.D., C.S., Xue.C., Q.T., J.Y., X.Z. and H.Y.; writing—original draft preparation, A.W., W.D., C.B. and H.B.A.; writing—review and editing, A.W., X.T. and Y.Y.; supervision, A.W., H.B.A., X.T. and Y.Y.; project administration, A.W., X.Z., H.Y. C.B. and X.T.; funding acquisition, A.W., H.Y., X.T. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
We acknowledge the administrative, technical and logistical support provided by Wenzhou Municipal Key Lab for Applied Biomedical and Biopharmaceutical Informatics and Zhejiang Bioinformatics International Science and Technology Cooperation Center, headed by Y.Y.
Funding
Wenzhou-Kean University Student Partnering with Faculty (SpF) research program (SpF2021002, SpS2021029 and WKU201920009) awarded to A.W. and H.Y.; Wenzhou Municipal Key Lab for Applied Biomedical and Biopharmaceutical Informatics (WB20211227000125); Zhejiang Bioinformatics International Science and Technology Cooperation Center (WB20210429000008) headed by Y.Y.
Conflict of interest
The authors declare that there is no conflict of interest.



