-
PDF
- Split View
-
Views
-
Cite
Cite
Lisa Langnickel, Johannes Darms, Katharina Heldt, Denise Ducks, Juliane Fluck, Continuous development of the semantic search engine preVIEW: from COVID-19 to long COVID, Database, Volume 2022, 2022, baac048, https://doi.org/10.1093/database/baac048
Close - Share Icon Share
Abstract
preVIEW is a freely available semantic search engine for Coronavirus disease (COVID-19)-related preprint publications. Currently, it contains >43 800 documents indexed with >4000 semantic concepts, annotated automatically. During the last 2 years, the dynamic situation of the corona crisis has demanded dynamic development. Whereas new semantic concepts have been added over time—such as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of interest—the service has been also extended with several features improving the usability and user friendliness. Most importantly, the user is now able to give feedback on detected semantic concepts, i.e. a user can mark annotations as true positives or false positives. In addition, we expanded our methods to construct search queries. The presented version of preVIEW also includes links to the peer-reviewed journal articles, if available. With the described system, we participated in the BioCreative VII interactive text-mining track and retrieved promising user-in-the-loop feedback. Additionally, as the occurrence of long-term symptoms after an infection with the virus SARS-CoV-2—called long COVID—is getting more and more attention, we have recently developed and incorporated a long COVID classifier based on state-of-the-art methods and manually curated data by experts. The service is freely accessible under https://preview.zbmed.de
Introduction
The rapid spread and social and economic impacts of the COVID-19 pandemic increased the need for the scientific community to understand the virus-induced disease and to provide means to elevate the impact. Hence, the need to share—even preliminary—research results with peers increased. Since the well-established peer-reviewed publication process takes time, the importance of preprint servers to share results without a peer-review process increased. In 2020, between 17% and 30% of the total COVID-19 research papers were published as preprints (1). Since those articles are not peer-reviewed they are also missing a peer-reviewed classification with meaningful keywords that are useful to find relevant articles. In addition, several preprint servers with partly overlapping domains exist, which makes it cumbersome to find relevant literature.
To facilitate the central access to these literature resources and enable semantic search, we have established a service, preVIEW COVID-19, that aggregates and combines the content from different preprint servers and enriches the articles with semantic information through an advanced, natural language processing (NLP)-based workflow. As a result, a one-stop-shop for COVID-19-related preprint publications with semantic search functionality is available. The service can be freely accessed via https://preview.zbmed.de and currently contains >43 800 preprints from seven different preprint servers. The system and text-mining components are described in detail in (2) and (3).
Since the initial release of our service in April 2020, we continuously added more data sources (preprint servers) and update the data daily. Further, we added new requested features to the system. In this work, we describe—next to a general system overview—the new features that we integrated based on requests of the BioCreative VII interactive text-mining (IAT) track and results are disseminated within this article. BioCreative is a community effort to evaluate and develop information extraction systems and takes place regularly since 2003 (https://biocreative.bioinformatics.udel.edu/). The IAT track was first established in 2011 to give system developers end-user feedback on their provided services (4). The last IAT track focused on systems that include COVID-19-related text-mining components that are evaluated by users performing both predefined and free tasks.
In addition, we have also continued to integrate new features after participating in the BioCreative VII IAT track. In particular, the occurrence of long-term symptoms after an infection with the virus SARS-CoV-2 is especially relevant for public health institutes such as the Robert Koch Institute (RKI) (https://www.rki.de/EN/Home/). The long COVID data basis is still small, and it is of great importance to collect all available data in order to find evidence and draw new conclusions. Currently, complex search queries are being developed by information specialists to find new relevant information in publications. There is no established terminology for long COVID descriptions and new information arise over time—as a consequence, search terms need to be readjusted continuously. Since first expert annotated data are available, the next logical step is to apply promising state-of-the-art algorithms, such as transformer-based models like Bidirectional Encoder Representations from Transformers (BERT) (5) for classification tasks. Those classification approaches should also be able to generalize and to predict on new data.
In the current work, we describe the new features developed in accordance to the IAT track and the corresponding results from the user study. In addition, the training of a document classifier based on the manually curated data for long-COVID-related articles and the integration of the classification results into our semantic search engine preVIEW are presented.
System Description
General System Description
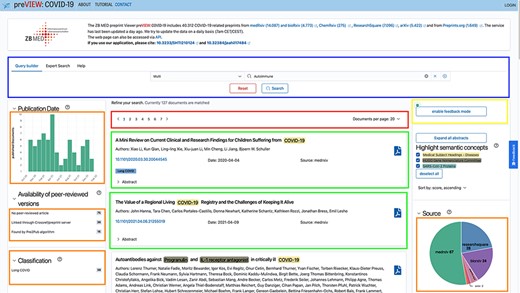
The service preVIEW COVID-19 can be accessed as a user via a web browser or programmatically via an application programming interface (API). The application is developed as an integrative single-page application. In Figure 1, the landing page is shown. On the top (blue box), a search query can be constructed to refine the result list of documents (green box). Additionally, the search result set can be further refined by using the facets provided on both sides of the screen (orange boxes): These include filter functions, such as displaying all preprints that contain at least one disease mention or adding specific disease concepts to your search query; for each of the four entity classes (diseases, human genes and proteins, and SARS-CoV-2-specific proteins and variants of concern), the five most frequently occurring concepts for the current subcorpus are displayed on the left. Clickable charts can be used to directly select a specific time period or preprint source (e.g. medRxiv). The search results can be exported in Bibtex or EndNote formats. Additionally, abstracts can be shown or hidden, and semantic concept annotation can be optionally highlighted. Semantic concepts are recognized by rule-based and machine-learning-based methods for disease concepts (mapped to medical subject headings), human genes (mapped to HumanGenome Nomenclature Committee), SARS-CoV-2 proteins (mapped to UniProt entries) and virus variants (retrieved from (6)).
New Features in the Context of BioCreative IAT Track 2021
We implemented new methods to link preprints to their corresponding journal articles, if available. The retrieval of this information and the integration into preVIEW are described in (7), here we give a short summary of the functionality: A filter to restrict the search to articles that have also been published in peer-reviewed journals is available as a facet in the upper left corner (orange box). The user can select all corresponding articles or select only specific sources from which we obtained the information: directly from the preprint servers, from Crossref or retrieved via an NLP-based algorithm Pre2Pub. By request of the organizers, we added the corresponding link to each preprint, if available.
On top right (yellow box), the feedback mode, one of our newly introduced features, can be enabled. When the mode is enabled, the semantic annotations are expanded with two icons (thumbs up and down) on the right side of every annotation. By clicking on an icon, the user sends feedback about the annotation to the system: Thumbs up indicating a correct annotation—i.e., true positive and thumbs down an incorrect (false-positive) one. The feature can be seen in Figure 2. For each feedback, we store a set of properties [document id, offset-begin, offset-end, concept internationalized resource identifier (IRI), feedback type (either thumbs up or down), reported time and user id, if the user is authenticated]. The tuple is stored within a mongodb database system. No IP addresses and other user-identifiable information is stored within the system as it is not necessary for our use case and corresponds to a required data economy of General Data Protection Regulation (GDPR) (8). We envision that this crowd-sourced information can be used to extend our training data and in such a way improve our text-mining components.

Screenshot of a single document with the feedback mode enabled (thumbs up/down).
In addition, we expanded our search functionality by an ‘expert search’. The user can now switch between the ‘query builder’ and the ‘expert search’ (see Figures 3 and 4). While the user can expand the search query by clicking buttons and selecting the data field from a list when using the Query Builder, the Expert Search allows the user to edit the query manually. The user can also first use the Query Builder—with the benefit of auto-completion when searching for a semantic concept—and then switch to the Expert Search to change the logical concatenation of different search terms.
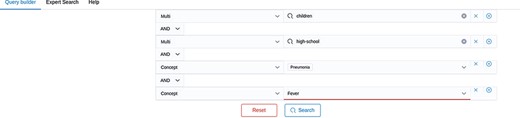
Screenshot of the Query Builder. Search terms can be added via drop-down lists.
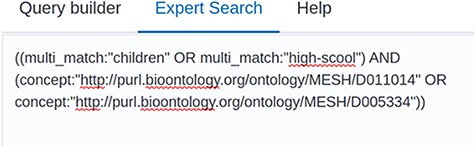
Screenshot of the Expert Search. The raw query string can be seen here and can be manually edited.
Long COVID Classification
In the following, we describe the data and methods used to train and evaluate the long-COVID-based document classifier.
Data
Long-COVID-related articles have been manually collected by information specialists. As a certain amount of data are needed to train a deep-learning-based model, data from two different sources have been merged in case of positive examples. To get further negative examples, we used a third resource.
The first subset was provided by information specialists from the RKI and has been collected in the following way: In the last two weeks of April 2021, the database PubMed was searched using the following search string:
(‘post-acute COVID-19 syndrome’[Supplementary Concept] OR ‘post-acute COVID-19 syndrome’[All Fields] OR ‘post covid*’[All Fields] OR ‘post covid*’[Title/Abstract] OR ‘long covid*’[All Fields] OR ‘long covid*’[Title/Abstract])
At that time, 1,644 articles were retrieved, but most of the papers focused on longitudinal studies related to COVID-19 in general. In collaboration with RKI’s scientists, it has been decided to combine the search string with filters for ‘clinical trials’, ‘randomized controlled trials’, ‘reviews’ and ‘systematic reviews’—resulting in more precise results focusing on post-COVID-19 Condition. Hence, a total of 237 papers were received with the adapted search query:
(‘post-acute COVID-19 syndrome’[Supplementary Concept] OR ‘post-acute COVID-19 syndrome’[All Fields] OR ‘post covid*’[All Fields] OR ‘post covid*’[Title/Abstract] OR ‘long covid*’[All Fields] OR ‘long covid*’[Title/Abstract]) AND (clinicaltrial[Filter] OR randomizedcontrolledtrial[Filter] OR review[Filter] OR systematicreview[Filter])
Moreover, the search engine LitCovid (9, 10) was queried with the search terms ‘long-covid’ OR ‘long covid’ (https://www.ncbi.nlm.nih.gov/research/coronavirus/docsum?text=%22long-covid%22%20OR%20%22long%20covid%22) and further 228 results were received. In addition, the in-house EndNote database provided by the RKI library (11) has been manually screened for the following search terms: ‘long-COVID’, ‘long-haul COVID’, ‘post-acute COVID syndrome’, ‘persistent COVID-19’, ‘post-acute COVID19 syndrome’, ‘long hauler COVID’, ‘long COVID’, ‘post-acute sequelae of SARS-CoV-2 infection’, ‘long haul COVID’ and ‘chronic COVID syndrome’. We also searched the World Health Organization literature hub for COVID-19 (12) with the search terms ‘long-covid’ and ‘long covid’ (https://search.bvsalud.org/global-literature-on-novel-coronavirus-2019-ncov/?output=site&lang=en&from=0&sort=&format=summary&count=20&fb=&page=1&skfp=&index=tw&q=%22long+covid%22+OR+long-covid&search_form_submit=). We received 230 results at the time. The references were pulled from the following databases: MEDLINE (106), medRxiv (40), Web of Science (13), ProQuest Central (12), International Clinical Trials Registry Platform (ICTRP) (9), PubMed (9), Scopus (8), Academic Search Complete (6), EMBASE (5), ScienceDirect (5), CINAHL (4), Wiley (4), PubMed Central (PMC) (3), bioRxiv (3), Multidisciplinary Digital Publishing Institute (MDPI) (2) and Africa Wide Information (1). RKI’s scientists provided also a few papers from other experts, networks or institutions. We removed duplicates by using the EndNote Citation manager and all results were manually screened by two experts and further labeled accordingly. The long-COVID-relevant papers are selected from the results and are part of the first training set. The papers that were not marked as related to long COVID are part of the negative examples. Thereby, we included only those references where an abstract was available using the PubMed API via the E-utilities (13). This resulted in 183 positive and 283 negative examples.
The second subset is retrieved from the ‘Long covid research library’ released by Pandemic-Aid Networks, who also manually collect ‘important papers that have been published on Long Covid’ (14). We retrieved 162 articles on 4 January 2022. As these are all positive examples, we needed further negative examples to have a balanced training dataset. Therefore, we used again the database LitCovid, where in the meantime long COVID labels were integrated with a human-in-the-loop machine learning approach (15), and filtered for non-long-COVID articles (query: NOT e_condition:LongCovid) (https://www.ncbi.nlm.nih.gov/research/coronavirus/docsum?text=NOT%20e_condition:LongCovid) and retrieved further 62 articles. The datasets have been merged, shuffled randomly and split into training, development and test sets. An overview about the data can be seen in Table 1.
| . | Training . | Development . | Test . | Total . |
|---|---|---|---|---|
| Positive examples | 215 | 76 | 70 | 345 |
| Negative examples | 199 | 62 | 68 | 345 |
| Total | 414 | 138 | 138 | 690 |
| . | Training . | Development . | Test . | Total . |
|---|---|---|---|---|
| Positive examples | 215 | 76 | 70 | 345 |
| Negative examples | 199 | 62 | 68 | 345 |
| Total | 414 | 138 | 138 | 690 |
| . | Training . | Development . | Test . | Total . |
|---|---|---|---|---|
| Positive examples | 215 | 76 | 70 | 345 |
| Negative examples | 199 | 62 | 68 | 345 |
| Total | 414 | 138 | 138 | 690 |
| . | Training . | Development . | Test . | Total . |
|---|---|---|---|---|
| Positive examples | 215 | 76 | 70 | 345 |
| Negative examples | 199 | 62 | 68 | 345 |
| Total | 414 | 138 | 138 | 690 |
Document Classification
For document classification, we fine-tuned the pre-trained BERT model ‘bert-base-cased’ (5) from Huggingface using the transformers library (16). We performed hyperparameter optimization using the development set for different batch sizes (8, 16, 32), learning rates (|$1e-5$|, |$3e-5$|, |$5e-5$|) and numbers of epochs (1–10). Maximum sequence length was set to 512 in all cases. For training the final model, both the development set and training set were combined. The used parameters can be seen in Table 2. Both data and model have been pushed to Huggingface and can be accessed under https://huggingface.co/datasets/llangnickel/long-covid-classification-data and https://huggingface.co/llangnickel/long-covid-classification, respectively.
| Parameter . | Value . |
|---|---|
| Learning rate | |$3e-5$| |
| Batch size | 16 |
| Number of epochs | 4 |
| Sequence length | 512 |
| Parameter . | Value . |
|---|---|
| Learning rate | |$3e-5$| |
| Batch size | 16 |
| Number of epochs | 4 |
| Sequence length | 512 |
| Parameter . | Value . |
|---|---|
| Learning rate | |$3e-5$| |
| Batch size | 16 |
| Number of epochs | 4 |
| Sequence length | 512 |
| Parameter . | Value . |
|---|---|
| Learning rate | |$3e-5$| |
| Batch size | 16 |
| Number of epochs | 4 |
| Sequence length | 512 |
Results
In the following, we will first describe the current preVIEW version with its new features. Afterward, the results of the usability tests are described in detail.
Description of the new preVIEW version
The current version of preVIEW is actively used with over 500 unique visitors per month and over 1500 requests per day; usage also increased during the challenge. Feedback received via mail or via the website itself indicates an active community using the service. For taking part in the challenge, three new features were requested by the organizers: first, the implementation of a feedback button for the provided annotations. Second, the link to a corresponding peer-reviewed article, if available. And finally, we extended the search functionalities for building queries.
PreVIEW users seem to accept the new feedback feature well, about 2 months after the release of the annotation feedback tool, we received already 110 feedback items from users not involved in the project development.
The second new feature provides links and filter function for corresponding peer-reviewed articles. From the current content, (out of more than 37,000 preprint articles), 7921 (around 21%) of all listed preprints are also published in a peer-reviewed journal. Thereof, 4232 articles have been detected only automatically. Finally, the search bar now contains three different tabs, which are the Query Builder, the Expert Search and Help. An example of a query built with the query builder can be seen in Figure 3. Here, two terms—high school or children—were searched together with two concepts—fever or pneumonia. In order to build the correct logical concatenation, in this example combining two OR-blocks by an AND, the user can switch to the Expert Search and change the parentheses (see Figure 4). In the Expert Search, for concepts the IRI is shown instead of the preferred label.
Results of the Usability Testing
Through participation in the BioCreative VII IAT Challenge, we received formal user feedback from eight individuals. The survey contains a total of 28 items, comprising a combination of free-text questions and ordinal questions. The first three questions are about the background and experience of the users; detailed results are summarized in Figure 5. Except for one user who is doing experimental work, all our test users indicated that they have a background in computer science and/or biocuration. It was also indicated that all participants were from the academic or government sectors and that the majority (i.e. 75%) is also actively involved in research related to COVID-19.
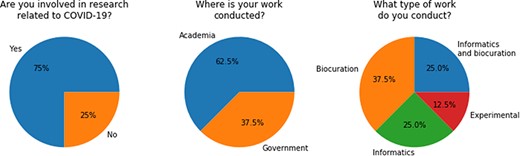
Questions about background and experiences as answered by the participants. The absolute number of persons is shown in brackets.
In questions 3–15, summarized in Table 3, the users were basically asked to describe what they did by giving examples of their tried queries and functionalities when they used the system. In addition, questions about things that work well or—in contrast—possible improvements were described, also in comparison to other systems already known by the users. Concerning the question ‘What thing(s) did you like best about the system?’ answers concerning both the implementation (e.g. ‘Everything is intuitive and requires minimal if no learning curve at all.’, ‘Retrieval was very fast and accurate’ and ‘Very user friendly’) and the purpose of the service itself (e.g. ‘Have access to preprints from different sources in one platform’ and ‘Having a source which collects COVID-19 related preprints from other preprint sources.’) were given. Seven out of eight users also found that the data/results were provided in an easy/useful format. However, the free-text responses also indicate that further improvements are needed. Some errors were found and some ideas and wishes were expressed: One feature is the desire for the ability to save and reuse searches, analogous to the functionality at PubMed. Furthermore, highlighting of results in free-text searches was requested.
| No . | Question . |
|---|---|
| 3 | Provide examples of queries you tried when exploring on your own. |
| 4 | Provide examples of functionalities tried when exploring on your own. |
| 5 | What thing(s) did you like best about this system? |
| 6 | What would you recommend to improve the system? If possible, list specific features you would like to see added. |
| 7 | Did you find any bottleneck while navigating the system? |
| 8 | If yes, please explain |
| 9 | Were the data/results provided to you in an easy/useful format? |
| 10 | Any suggestion for improvement? |
| 11 | Were the outputs useful? |
| 12 | Any suggestion for improvement? |
| 13 | Are there applications or kinds of work that you think this system would be useful for? If so, please list or describe briefly. |
| 14 | Before participating in this BioCreative activity, were you aware or have you used any similar system? |
| 15 | If yes, could you indicate what system/tool have you used and how it compares to this one? |
| No . | Question . |
|---|---|
| 3 | Provide examples of queries you tried when exploring on your own. |
| 4 | Provide examples of functionalities tried when exploring on your own. |
| 5 | What thing(s) did you like best about this system? |
| 6 | What would you recommend to improve the system? If possible, list specific features you would like to see added. |
| 7 | Did you find any bottleneck while navigating the system? |
| 8 | If yes, please explain |
| 9 | Were the data/results provided to you in an easy/useful format? |
| 10 | Any suggestion for improvement? |
| 11 | Were the outputs useful? |
| 12 | Any suggestion for improvement? |
| 13 | Are there applications or kinds of work that you think this system would be useful for? If so, please list or describe briefly. |
| 14 | Before participating in this BioCreative activity, were you aware or have you used any similar system? |
| 15 | If yes, could you indicate what system/tool have you used and how it compares to this one? |
| No . | Question . |
|---|---|
| 3 | Provide examples of queries you tried when exploring on your own. |
| 4 | Provide examples of functionalities tried when exploring on your own. |
| 5 | What thing(s) did you like best about this system? |
| 6 | What would you recommend to improve the system? If possible, list specific features you would like to see added. |
| 7 | Did you find any bottleneck while navigating the system? |
| 8 | If yes, please explain |
| 9 | Were the data/results provided to you in an easy/useful format? |
| 10 | Any suggestion for improvement? |
| 11 | Were the outputs useful? |
| 12 | Any suggestion for improvement? |
| 13 | Are there applications or kinds of work that you think this system would be useful for? If so, please list or describe briefly. |
| 14 | Before participating in this BioCreative activity, were you aware or have you used any similar system? |
| 15 | If yes, could you indicate what system/tool have you used and how it compares to this one? |
| No . | Question . |
|---|---|
| 3 | Provide examples of queries you tried when exploring on your own. |
| 4 | Provide examples of functionalities tried when exploring on your own. |
| 5 | What thing(s) did you like best about this system? |
| 6 | What would you recommend to improve the system? If possible, list specific features you would like to see added. |
| 7 | Did you find any bottleneck while navigating the system? |
| 8 | If yes, please explain |
| 9 | Were the data/results provided to you in an easy/useful format? |
| 10 | Any suggestion for improvement? |
| 11 | Were the outputs useful? |
| 12 | Any suggestion for improvement? |
| 13 | Are there applications or kinds of work that you think this system would be useful for? If so, please list or describe briefly. |
| 14 | Before participating in this BioCreative activity, were you aware or have you used any similar system? |
| 15 | If yes, could you indicate what system/tool have you used and how it compares to this one? |
Additionally, a bug was found in the query builder related to NOT queries. Furthermore, false-positive and false-negative text-mining annotations were criticized. Semantic annotation of compounds and the ability to provide custom dictionaries for annotation were also requested.
Questions 16–28 are ordinal questions. Out of these ordinal questions, all except one are on a scale of one to five, with one representing a negative perception and five representing strong agreement. The last question (no. 28) has an ordinal scale of one to ten. For each question, we provide results as the average and median values, shown in Table 4.
Overview of survey questions [*mean and average values for ordinal question of the user survey. Ranges: [1,5] (strong disagree, strong agree). **[1,10] (not likely at all, extremely likely)]
| No. . | Question* . | Average . | Median . |
|---|---|---|---|
| 16 | I think that I would like to use this system frequently | 3.38 | 4 |
| 17 | I found the system unnecessarily complex | 1.88 | 2 |
| 18 | I thought the system was easy to use | 4.5 | 5 |
| 19 | I think I would need support from the developer to be able to use this system | 1.13 | 1 |
| 20 | I found the various functions of the system well integrated | 3.88 | 4 |
| 21 | I thought there was too much inconsistency in this system | 1.88 | 1.5 |
| 22 | I would imagine that most people would learn to use this system very quickly | 4.75 | 5 |
| 23 | I found the system very cumbersome to use | 1.38 | 1 |
| 24 | The system has met my expectations | 4.25 | 4.5 |
| 25 | I felt very confident using the system | 4.5 | 4.5 |
| 26 | I needed to learn a lot of things before I could get going with the system | 1.75 | 1 |
| 27 | Please rate your overall impression with the system | 4.13 | 4 |
| 28** | How likely is that you would recommend this system to a colleague performing COVID-19 related research? | 7.63 | 8.5 |
| No. . | Question* . | Average . | Median . |
|---|---|---|---|
| 16 | I think that I would like to use this system frequently | 3.38 | 4 |
| 17 | I found the system unnecessarily complex | 1.88 | 2 |
| 18 | I thought the system was easy to use | 4.5 | 5 |
| 19 | I think I would need support from the developer to be able to use this system | 1.13 | 1 |
| 20 | I found the various functions of the system well integrated | 3.88 | 4 |
| 21 | I thought there was too much inconsistency in this system | 1.88 | 1.5 |
| 22 | I would imagine that most people would learn to use this system very quickly | 4.75 | 5 |
| 23 | I found the system very cumbersome to use | 1.38 | 1 |
| 24 | The system has met my expectations | 4.25 | 4.5 |
| 25 | I felt very confident using the system | 4.5 | 4.5 |
| 26 | I needed to learn a lot of things before I could get going with the system | 1.75 | 1 |
| 27 | Please rate your overall impression with the system | 4.13 | 4 |
| 28** | How likely is that you would recommend this system to a colleague performing COVID-19 related research? | 7.63 | 8.5 |
Overview of survey questions [*mean and average values for ordinal question of the user survey. Ranges: [1,5] (strong disagree, strong agree). **[1,10] (not likely at all, extremely likely)]
| No. . | Question* . | Average . | Median . |
|---|---|---|---|
| 16 | I think that I would like to use this system frequently | 3.38 | 4 |
| 17 | I found the system unnecessarily complex | 1.88 | 2 |
| 18 | I thought the system was easy to use | 4.5 | 5 |
| 19 | I think I would need support from the developer to be able to use this system | 1.13 | 1 |
| 20 | I found the various functions of the system well integrated | 3.88 | 4 |
| 21 | I thought there was too much inconsistency in this system | 1.88 | 1.5 |
| 22 | I would imagine that most people would learn to use this system very quickly | 4.75 | 5 |
| 23 | I found the system very cumbersome to use | 1.38 | 1 |
| 24 | The system has met my expectations | 4.25 | 4.5 |
| 25 | I felt very confident using the system | 4.5 | 4.5 |
| 26 | I needed to learn a lot of things before I could get going with the system | 1.75 | 1 |
| 27 | Please rate your overall impression with the system | 4.13 | 4 |
| 28** | How likely is that you would recommend this system to a colleague performing COVID-19 related research? | 7.63 | 8.5 |
| No. . | Question* . | Average . | Median . |
|---|---|---|---|
| 16 | I think that I would like to use this system frequently | 3.38 | 4 |
| 17 | I found the system unnecessarily complex | 1.88 | 2 |
| 18 | I thought the system was easy to use | 4.5 | 5 |
| 19 | I think I would need support from the developer to be able to use this system | 1.13 | 1 |
| 20 | I found the various functions of the system well integrated | 3.88 | 4 |
| 21 | I thought there was too much inconsistency in this system | 1.88 | 1.5 |
| 22 | I would imagine that most people would learn to use this system very quickly | 4.75 | 5 |
| 23 | I found the system very cumbersome to use | 1.38 | 1 |
| 24 | The system has met my expectations | 4.25 | 4.5 |
| 25 | I felt very confident using the system | 4.5 | 4.5 |
| 26 | I needed to learn a lot of things before I could get going with the system | 1.75 | 1 |
| 27 | Please rate your overall impression with the system | 4.13 | 4 |
| 28** | How likely is that you would recommend this system to a colleague performing COVID-19 related research? | 7.63 | 8.5 |
Overall, the mean values show a general satisfaction with the system. For example, for the statement ‘I think that I would like to use this system frequently’, there is an average of 3.38 and a median of 4. A low average of about one for the statement ‘I think I would need support from a developer to be able to use the system’ indicates a high level of usability of the system. This is confirmed by Statement 25 ‘I felt very confident using the system’ where a median of 4.5 is achieved. The answers to the last two questions ‘Please rate your overall impression with the system’ and ‘How likely is that you would recommend this system to a colleague performing COVID-19 related research’ indicate a good overall all performance of the system with the mean values 4.13 (out of 5) and 7.63 (out of ten), respectively.
Performance and Integration of Long COVID Classifier
The trained long COVID classifier has been evaluated on the independent test set. It reaches a high performance with an F1-score of 91.18%, resulting from equally high precision and recall.
A detailed error analysis revealed that, among the false-positive predictions, most of the articles describe severe COVID-19 cases where patients suffer from chronic diseases. Another false-positive example talks about long-term implications not related to the COVID-19 infection itself but to the treatment due to the infection. For false-negative examples, at first glance, there are no similarities to be seen. However, several among them are reviews or meta-analyses.
The long COVID classifier is integrated into the search engine preVIEW and reveals that, out of 40 312 COVID-19-related preprints, 3132 are classified as long-COVID-related articles—which makes up ~8%. As can be seen in Figure 6, a filter option has been implemented with which it is easily possible to select only long-COVID-related articles. In addition, it is indicated on each article with a blue long COVID label. This implementation allows for easy extension if further classifications will be added.

Discussion
Our service preVIEW has been developed as a first prototype right at the beginning of the COVID-19 pandemic in order to support researchers in quickly finding relevant information. Based on user feedback, we continuously improved the system and added new semantic concepts and new features.
In order to get a broader and more structured user feedback, also from users we are not in direct contact with, we participated in the IAT challenge—which has certainly made the services more popular and attracted new users. Through the structured feedback and free-text responses, we have gained a deeper insight into the users’ requirements which will guide the direction of our future developments. The structured feedback from eight participants and the descriptive statistics derived from it show an application that meets current needs and would be overall recommended (7.63)—with two participants being very likely to recommend the system to others (=10) and one would not do so at all (=1). This suggests that more participants are needed for meaningful statistics from the survey. In general, we got the feedback that the basic design and implementation is very intuitive and easy-to-use. However, several features for improvement of the search were requested, such as highlighting the query match or allowing the users to upload their own terminology for semantic retrieval of a specific field of research where no standardized vocabulary exists.
The RKI information specialists requested a further update of our system—a filter function for long-COVID-related documents. These long-term effects of the COVID-19 disease are of high overall relevance for public health. There are already promising state-of-the-art methods for document classification that offer enormous advantages over classical retrieval with Boolean queries, especially for non-terminology-indexed data (such as Medical Subject Headings (MeSH) indexing of PubMed) and particularly in terms of generalizability. However, the performance of these methods depends crucially on the amount and quality of labeled training data. The training data used here was mainly manually curated by information specialists in a time-consuming process. Based on a dataset of 700 abstracts, we were able to train a powerful classifier that achieves an F1-score of 91.18% in the independent test set. Integrating the classification results into the search engine preVIEW showed that about 8% of the articles are relevant to long COVID. With the feedback mode already implemented in preVIEW, we can easily extend the training dataset to enable further improvements to the current classification system.
The quickly changing, dynamic situation of the current corona crisis has demanded a dynamic development. Therefore, we continuously improved our semantic search engine and added new features and semantic concepts. Getting user feedback is valuable and indispensable for developing a user-friendly service. Through our participation in the BioCreative IAT challenge and the direct cooperation with information specialists, we could continuously improve our system based on user needs. In general, the feedback is very valuable and—since the role of preprints is increasing enormously—can pave the way for new features and/or new semantic search systems, even concerning a more general use case (apart from COVID-19). Therefore, ZB MED will use the feedback to elaborate on the future developments concerning their digital information services.
Conclusion
Through participation in the BioCreative IAT challenge, we got valuable and positive feedback for the usability of our semantic search engine preVIEW COVID-19. As the importance of preprints is increasing not only for the COVID-19-related research, the establishment of a permanent service at ZB MED for all preprints will be future work.
Acknowledgement
We would like to thank the BioCreative IAT track organizers and testers for their valuable feedback! Moreover, we would like to thank Roman Baum (ZB MED—Information Centre for Life Sciences) and Julia Sasse (ZB MED—Information Centre for Life Sciences) for their support in software development.
Funding
NFDI4Health (National Research Data Infrastructure for Personal Health Data) Task Force COVID-19 (www.nfdi4health.de); Deutsche Forschungsgemeinschaft or German Research Foundation (451265285).
Competing interests
There is no competing interest.
Author contributions statement
L.L. and J.D. developed the software and wrote and reviewed the manuscript. K.H. and D.D. provided the manually curated training data and also reviewed the manuscript. J.F. reviewed the manuscript and supervised the project.



