-
PDF
- Split View
-
Views
-
Cite
Cite
Rhoda J. Kinsella, Andreas Kähäri, Syed Haider, Jorge Zamora, Glenn Proctor, Giulietta Spudich, Jeff Almeida-King, Daniel Staines, Paul Derwent, Arnaud Kerhornou, Paul Kersey, Paul Flicek, Ensembl BioMarts: a hub for data retrieval across taxonomic space, Database, Volume 2011, 2011, bar030, https://doi.org/10.1093/database/bar030
Close - Share Icon Share
Abstract
For a number of years the BioMart data warehousing system has proven to be a valuable resource for scientists seeking a fast and versatile means of accessing the growing volume of genomic data provided by the Ensembl project. The launch of the Ensembl Genomes project in 2009 complemented the Ensembl project by utilizing the same visualization, interactive and programming tools to provide users with a means for accessing genome data from a further five domains: protists, bacteria, metazoa, plants and fungi. The Ensembl and Ensembl Genomes BioMarts provide a point of access to the high-quality gene annotation, variation data, functional and regulatory annotation and evolutionary relationships from genomes spanning the taxonomic space. This article aims to give a comprehensive overview of the Ensembl and Ensembl Genomes BioMarts as well as some useful examples and a description of current data content and future objectives.
Database URLs:http://www.ensembl.org/biomart/martview/; http://metazoa.ensembl.org/biomart/martview/; http://plants.ensembl.org/biomart/martview/; http://protists.ensembl.org/biomart/martview/; http://fungi.ensembl.org/biomart/martview/; http://bacteria.ensembl.org/biomart/martview/
Project description
The Ensembl project (http://www.ensembl.org) was launched in 2000 and is a joint effort by the European Bioinformatics Institute (EBI) and the Wellcome Trust Sanger Institute (WTSI). Ensembl aims to provide high-quality genomic resources including gene annotations, multiple sequence alignments, whole-genome variation data and other information valuable for reuse by the community in a wide variety of research contexts (1).
As of release 61 (February 2011), 56 species are supported in Ensembl. The project focuses its support on chordate species and particularly on human genome resources and those of key model organisms such as mouse, rat and zebrafish. Ensembl also includes three non-chordate species because of their historical use as models for basic biological process. Four of the 56 supported species are in a pre-release state and can be viewed at http://pre.ensembl.org. The remaining 52 species all include comprehensive, evidence-based gene annotations and assignments of gene homology relationships. A smaller number of species include additional genomic data resources, largely chosen as a result of data availability and collaboration with species-specific or targeted resources. For example, Ensembl variation data resources include those in dbSNP (2) as well as variation data created by the project in the context of genome analysis (3). Close collaboration with other projects at the EBI including InterPro (4), the Database of Genomic Variants archive (DGVa) (5) and HGNC (6) ensures that Ensembl resources are integrated and available through other important bioinformatics resources. Recently somatic mutation data from the Catalogue of Somatic Mutations in Cancer (COSMIC) (7) has been incorporated into the Ensembl variation database.
The Ensembl Genomes project (http://www.ensemblgenomes.org) is comprised of separate websites for five distinct domains of life: bacteria, fungi, protists, plants and invertebrate metazoa (8). This project utilizes the Ensembl tools to provide genome-centric resources for species spanning the taxonomic space. Since the project launch in 2009, this portal has increased the number of genomes it represents from 122 species (bacteria, metazoa and protists) to 313 species (Ensembl Genomes release 8) of non-vertebrate genomes. For many species, the annotation is produced through collaborative efforts with scientific communities specializing in a particular domain, supplemented by the import of other publicly available information, while data from other important species is imported from various public repositories.
Ensembl and Ensembl Genomes are totally open projects and encourage others to incorporate the Ensembl code into their projects as well as provide specific tools for comprehensive data analysis and mining of the Ensembl data resources. In addition to long standing data resources such as the Ensembl gene sets (9) and gene trees (10), Ensembl provides other resources such as up-to-date microarray annotations (11). Widely used tools include the Variant Effect Predictor (VEP) (12) and the Ensembl API (13). The Ensembl genome browser at http://www.ensembl.org (14) provides a comprehensive visualization for accessing and using Ensembl data. The Ensembl BioMart (15,24) provides a final method for data access and querying data. Since the formative years of the Ensembl project, the BioMart data management system has played an important part in providing access for the scientific community to the growing volume of genome data. Each of the five Ensembl Genomes portals also contains a BioMart for optimized querying of the data.
Data content
The Ensembl BioMarts are created using the database schemas and data generated by the various components of the Ensembl project. The Ensembl BioMarts are comprised of seven databases (three hidden and four visible). The four visible databases on the BioMart interface are: Ensembl Genes, Ensembl Variation, Ensembl Regulation and Vega. The three hidden BioMart databases contain supporting information for the visible databases including sequence data, ontology data and miscellaneous genomic features such as Encyclopedia of DNA Elements (ENCODE) (16) and karyotype data. The data in these three databases are accessed via the visible BioMart databases on the interface. Additional databases are integrated from the PRIDE (17) and Reactome (18,22) projects using the BioMart database federation technology. The gene-centric Ensembl Genes database as of Ensembl release 61 contains 52 fully supported species, the Ensembl Variation database contains variation-centric data for 18 species, the Ensembl Regulation feature-set-centric database contains data for three species and the Vega database contains manually annotated gene-centric data for three species (Table 1).
| Data set . | Description of data content . |
|---|---|
| Ensembl Genes 61 | Genes from 52 species with annotated external references, protein domains, multi species comparison (orthologs, possible orthologs and paralogs), variation (germline and somatic), regulation (probe set mapping for microarray platforms), gene ontology, expression (GNF/Atlas) and transcript splicing event data |
| Ensembl Variation 61 | Variation data for 18 species including human somatic mutation data from COSMIC (7), human structural variation, human phenotype, Genome Wide Association Studies (GWAS) and variation set data. Strain specific data is available for certain other species. |
| Ensembl Regulation 61 | Regulation data for human, mouse and Drosophila melanogaster (annotated, regulatory and external features) |
| Vega 41 | Manually curated genes for human, mouse and zebrafish by the HAVANA group at WTSI and displayed in the VEGA database (21) |
| Reactome | Manually curated and peer-reviewed pathways from the BioMart (22) at http://www.reactome.org/ cgi-bin/mart |
| PRIDE (EBI UK) | Proteomics data from the PRIDE PRoteomics IDEntifications (17) BioMart database at http://www.ebi.ac.uk/pride/prideMart.do |
| Data set . | Description of data content . |
|---|---|
| Ensembl Genes 61 | Genes from 52 species with annotated external references, protein domains, multi species comparison (orthologs, possible orthologs and paralogs), variation (germline and somatic), regulation (probe set mapping for microarray platforms), gene ontology, expression (GNF/Atlas) and transcript splicing event data |
| Ensembl Variation 61 | Variation data for 18 species including human somatic mutation data from COSMIC (7), human structural variation, human phenotype, Genome Wide Association Studies (GWAS) and variation set data. Strain specific data is available for certain other species. |
| Ensembl Regulation 61 | Regulation data for human, mouse and Drosophila melanogaster (annotated, regulatory and external features) |
| Vega 41 | Manually curated genes for human, mouse and zebrafish by the HAVANA group at WTSI and displayed in the VEGA database (21) |
| Reactome | Manually curated and peer-reviewed pathways from the BioMart (22) at http://www.reactome.org/ cgi-bin/mart |
| PRIDE (EBI UK) | Proteomics data from the PRIDE PRoteomics IDEntifications (17) BioMart database at http://www.ebi.ac.uk/pride/prideMart.do |
| Data set . | Description of data content . |
|---|---|
| Ensembl Genes 61 | Genes from 52 species with annotated external references, protein domains, multi species comparison (orthologs, possible orthologs and paralogs), variation (germline and somatic), regulation (probe set mapping for microarray platforms), gene ontology, expression (GNF/Atlas) and transcript splicing event data |
| Ensembl Variation 61 | Variation data for 18 species including human somatic mutation data from COSMIC (7), human structural variation, human phenotype, Genome Wide Association Studies (GWAS) and variation set data. Strain specific data is available for certain other species. |
| Ensembl Regulation 61 | Regulation data for human, mouse and Drosophila melanogaster (annotated, regulatory and external features) |
| Vega 41 | Manually curated genes for human, mouse and zebrafish by the HAVANA group at WTSI and displayed in the VEGA database (21) |
| Reactome | Manually curated and peer-reviewed pathways from the BioMart (22) at http://www.reactome.org/ cgi-bin/mart |
| PRIDE (EBI UK) | Proteomics data from the PRIDE PRoteomics IDEntifications (17) BioMart database at http://www.ebi.ac.uk/pride/prideMart.do |
| Data set . | Description of data content . |
|---|---|
| Ensembl Genes 61 | Genes from 52 species with annotated external references, protein domains, multi species comparison (orthologs, possible orthologs and paralogs), variation (germline and somatic), regulation (probe set mapping for microarray platforms), gene ontology, expression (GNF/Atlas) and transcript splicing event data |
| Ensembl Variation 61 | Variation data for 18 species including human somatic mutation data from COSMIC (7), human structural variation, human phenotype, Genome Wide Association Studies (GWAS) and variation set data. Strain specific data is available for certain other species. |
| Ensembl Regulation 61 | Regulation data for human, mouse and Drosophila melanogaster (annotated, regulatory and external features) |
| Vega 41 | Manually curated genes for human, mouse and zebrafish by the HAVANA group at WTSI and displayed in the VEGA database (21) |
| Reactome | Manually curated and peer-reviewed pathways from the BioMart (22) at http://www.reactome.org/ cgi-bin/mart |
| PRIDE (EBI UK) | Proteomics data from the PRIDE PRoteomics IDEntifications (17) BioMart database at http://www.ebi.ac.uk/pride/prideMart.do |
The Ensembl Genomes BioMarts are created using the BioMart database schemas generated by the Ensembl project and these are adapted to suit the specific requirements for each of the domains. A gene-centric database is available for each of the five domains and a variation-centric database is available for Protists, Fungi, Metazoa and Plants (Table 2).
Summary of data available at the Ensembl Genomes BioMarts as of Ensembl Genomes release 8
| Data set . | Description of data content . |
|---|---|
| Ensembl Bacteria 8 | 249 genomes across 10 different clades (Gene database) |
| Ensembl Protists 8 | 11 species including Plasmodium falciparum, Plasmodium knowlesi, Plasmodium vivax and three oomycete genomes (Gene database for all species and Variation database for one species) |
| Ensembl Fungi 8 | 13 species, including eight Aspergillus species, Neosartorya fischeri, Puccinia graminis f. sp. Tritici, Saccharomyces cerevisiae, Schizosaccharomyces pombe (Gene database for all species and Variation database for one species) |
| Ensembl Metazoa 8 | 30 species, including 12 Drosphila, five Caenorhabditis, Aedes aegypti and Apis mellifera (Gene database for all species and Variation database for two species) |
| Ensembl Plants 8 | 10 species, including Arabidopsis lyrata, Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, Oryza sativa indica group and Zea mays (Gene database for all species and Variation database for four species) |
| Data set . | Description of data content . |
|---|---|
| Ensembl Bacteria 8 | 249 genomes across 10 different clades (Gene database) |
| Ensembl Protists 8 | 11 species including Plasmodium falciparum, Plasmodium knowlesi, Plasmodium vivax and three oomycete genomes (Gene database for all species and Variation database for one species) |
| Ensembl Fungi 8 | 13 species, including eight Aspergillus species, Neosartorya fischeri, Puccinia graminis f. sp. Tritici, Saccharomyces cerevisiae, Schizosaccharomyces pombe (Gene database for all species and Variation database for one species) |
| Ensembl Metazoa 8 | 30 species, including 12 Drosphila, five Caenorhabditis, Aedes aegypti and Apis mellifera (Gene database for all species and Variation database for two species) |
| Ensembl Plants 8 | 10 species, including Arabidopsis lyrata, Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, Oryza sativa indica group and Zea mays (Gene database for all species and Variation database for four species) |
Summary of data available at the Ensembl Genomes BioMarts as of Ensembl Genomes release 8
| Data set . | Description of data content . |
|---|---|
| Ensembl Bacteria 8 | 249 genomes across 10 different clades (Gene database) |
| Ensembl Protists 8 | 11 species including Plasmodium falciparum, Plasmodium knowlesi, Plasmodium vivax and three oomycete genomes (Gene database for all species and Variation database for one species) |
| Ensembl Fungi 8 | 13 species, including eight Aspergillus species, Neosartorya fischeri, Puccinia graminis f. sp. Tritici, Saccharomyces cerevisiae, Schizosaccharomyces pombe (Gene database for all species and Variation database for one species) |
| Ensembl Metazoa 8 | 30 species, including 12 Drosphila, five Caenorhabditis, Aedes aegypti and Apis mellifera (Gene database for all species and Variation database for two species) |
| Ensembl Plants 8 | 10 species, including Arabidopsis lyrata, Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, Oryza sativa indica group and Zea mays (Gene database for all species and Variation database for four species) |
| Data set . | Description of data content . |
|---|---|
| Ensembl Bacteria 8 | 249 genomes across 10 different clades (Gene database) |
| Ensembl Protists 8 | 11 species including Plasmodium falciparum, Plasmodium knowlesi, Plasmodium vivax and three oomycete genomes (Gene database for all species and Variation database for one species) |
| Ensembl Fungi 8 | 13 species, including eight Aspergillus species, Neosartorya fischeri, Puccinia graminis f. sp. Tritici, Saccharomyces cerevisiae, Schizosaccharomyces pombe (Gene database for all species and Variation database for one species) |
| Ensembl Metazoa 8 | 30 species, including 12 Drosphila, five Caenorhabditis, Aedes aegypti and Apis mellifera (Gene database for all species and Variation database for two species) |
| Ensembl Plants 8 | 10 species, including Arabidopsis lyrata, Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, Oryza sativa indica group and Zea mays (Gene database for all species and Variation database for four species) |
The Ensembl BioMart tables are made available for download from the FTP site (ftp://ftp.ensembl.org/pub) for each release (e.g. Ensembl Genes 61 BioMart database available from ftp://ftp.ensembl.org/pub/release-61/mysql/ensembl_mart_61). Users can access the BioMarts by web interface, BioMart API, biomaRt package from bioconductor (19), SOAP based and RESTful webservices and by publicly available MySQL server offering direct access to the BioMart databases (http://www.ensembl.org/info/data/mysql.html). Help and documentation details are summarized in Table 3. The Ensembl and Ensembl Genomes BioMarts are also displayed on the main BioMart central portal http://www.biomart.org. Three Ensembl mirrors have been created to improve the website performance for users around the globe. These mirrors, located on the west and east coasts of the USA (http://uswest.ensembl.org, http://useast.ensembl.org) and in Asia (http://asia.ensembl.org) also contain the Ensembl BioMarts to facilitate more effective data access.
| Information resource . | URL or Email address . |
|---|---|
| Ensembl frequently asked questions | http://www.ensembl.org/Help/Faq |
| BioMart frequently asked questions | http://www.biomart.org/faqs.html |
| Tutorials | http://www.ensembl.org/info/website/tutorials |
| YouTube videos | http://www.youtube.com/user/EnsemblHelpdesk |
| Ensembl news containing information about updates to mart databases | http://www.ensembl.org/info/website/news |
| Ensembl Blog | http://www.ensembl.info |
| Ensembl archives containing archived BioMart databases | http://www.ensembl.org/info/website/archives |
| Ensembl helpdesk mailing list | helpdesk@ensembl.org |
| Ensembl Genomes helpdesk mailing list | helpdesk@ensemblgenomes.org |
| Ensembl Genomes portal website containing project information | http://www.ensemblgenomes.org |
| Information resource . | URL or Email address . |
|---|---|
| Ensembl frequently asked questions | http://www.ensembl.org/Help/Faq |
| BioMart frequently asked questions | http://www.biomart.org/faqs.html |
| Tutorials | http://www.ensembl.org/info/website/tutorials |
| YouTube videos | http://www.youtube.com/user/EnsemblHelpdesk |
| Ensembl news containing information about updates to mart databases | http://www.ensembl.org/info/website/news |
| Ensembl Blog | http://www.ensembl.info |
| Ensembl archives containing archived BioMart databases | http://www.ensembl.org/info/website/archives |
| Ensembl helpdesk mailing list | helpdesk@ensembl.org |
| Ensembl Genomes helpdesk mailing list | helpdesk@ensemblgenomes.org |
| Ensembl Genomes portal website containing project information | http://www.ensemblgenomes.org |
| Information resource . | URL or Email address . |
|---|---|
| Ensembl frequently asked questions | http://www.ensembl.org/Help/Faq |
| BioMart frequently asked questions | http://www.biomart.org/faqs.html |
| Tutorials | http://www.ensembl.org/info/website/tutorials |
| YouTube videos | http://www.youtube.com/user/EnsemblHelpdesk |
| Ensembl news containing information about updates to mart databases | http://www.ensembl.org/info/website/news |
| Ensembl Blog | http://www.ensembl.info |
| Ensembl archives containing archived BioMart databases | http://www.ensembl.org/info/website/archives |
| Ensembl helpdesk mailing list | helpdesk@ensembl.org |
| Ensembl Genomes helpdesk mailing list | helpdesk@ensemblgenomes.org |
| Ensembl Genomes portal website containing project information | http://www.ensemblgenomes.org |
| Information resource . | URL or Email address . |
|---|---|
| Ensembl frequently asked questions | http://www.ensembl.org/Help/Faq |
| BioMart frequently asked questions | http://www.biomart.org/faqs.html |
| Tutorials | http://www.ensembl.org/info/website/tutorials |
| YouTube videos | http://www.youtube.com/user/EnsemblHelpdesk |
| Ensembl news containing information about updates to mart databases | http://www.ensembl.org/info/website/news |
| Ensembl Blog | http://www.ensembl.info |
| Ensembl archives containing archived BioMart databases | http://www.ensembl.org/info/website/archives |
| Ensembl helpdesk mailing list | helpdesk@ensembl.org |
| Ensembl Genomes helpdesk mailing list | helpdesk@ensemblgenomes.org |
| Ensembl Genomes portal website containing project information | http://www.ensemblgenomes.org |
Query examples
To demonstrate the utility of the Ensembl and Ensembl Genomes BioMarts we present several biologically relevant queries that can be performed using available tools and interfaces.
Query #1: The G-protein coupled receptor domain (GPCR) has the InterPro ID of IPR000276. Find the human protein-coding genes in Ensembl that code for this domain, and investigate whether any of them are detectable with the Affy HuGene 1_0 st v1 array.
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | Gene type: protein_coding | Ensembl Gene ID |
| Limit to genes with these family or domain IDs: IPR000276 | Associated Gene Name | |
| Affy HuGene 1_0 st v1 |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | Gene type: protein_coding | Ensembl Gene ID |
| Limit to genes with these family or domain IDs: IPR000276 | Associated Gene Name | |
| Affy HuGene 1_0 st v1 |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | Gene type: protein_coding | Ensembl Gene ID |
| Limit to genes with these family or domain IDs: IPR000276 | Associated Gene Name | |
| Affy HuGene 1_0 st v1 |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | Gene type: protein_coding | Ensembl Gene ID |
| Limit to genes with these family or domain IDs: IPR000276 | Associated Gene Name | |
| Affy HuGene 1_0 st v1 |
The GPCR genes make up a large protein family that covers a wide range of functions. A scientist may already know the InterPro ID of the GPCR rhodopsin-like domain and wish to investigate how many Ensembl gene IDs code for this GPCR and whether these were detected using the Affy HuGene 1_0 st v1 array. To do this query, the user must select the protein_coding filter from the GENE filter section and filter with the known InterPro ID in the PROTEIN DOMAINS filter section. Attributes are selected from Features:GENE and Features:EXTERNAL sections (Figure 1).
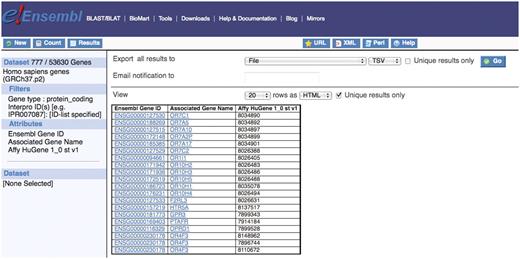
There are 777 Ensembl protein coding genes that code for the GPCR domain with InterPro ID (IPR000276) and that are detectable with the Affy HuGene 1_0 st v1 array 25.
Query #2: esv263 is the DGVa accession number of a structural variation from Redon et al. (20). What genomic region does this copy number variation span?
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Structural Variation | Limit to variants with these IDs: esv263 | Chromosome Name |
| Sequence region start (bp) | ||
| Sequence region end (bp) | ||
| Structural Variation Name | ||
| Structural Variation Description | ||
| Source Name |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Structural Variation | Limit to variants with these IDs: esv263 | Chromosome Name |
| Sequence region start (bp) | ||
| Sequence region end (bp) | ||
| Structural Variation Name | ||
| Structural Variation Description | ||
| Source Name |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Structural Variation | Limit to variants with these IDs: esv263 | Chromosome Name |
| Sequence region start (bp) | ||
| Sequence region end (bp) | ||
| Structural Variation Name | ||
| Structural Variation Description | ||
| Source Name |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Structural Variation | Limit to variants with these IDs: esv263 | Chromosome Name |
| Sequence region start (bp) | ||
| Sequence region end (bp) | ||
| Structural Variation Name | ||
| Structural Variation Description | ||
| Source Name |
Recent studies such as Redon et al. (20) have mapped copy number variations (CNV) in the human population. Redon et al. (20) studied 270 individuals from four populations whose DNA was screened for CNVs. Having read the article, a user may be interested in finding out more about a particular structural variation, such as the size of the genomic region that a particular structural variation spans (Figure 2). To do this query, the user must filter on the Structural Variation Name in the GENERAL STRUCTURAL VARIATION FILTERS and the attributes can be selected from the STRUCTURAL VARIATION attribute section.

The esv263 structural variation from DGVa occurs between 16 265 092 and 16 446 378 bp on chromosome 12.
Query #3: Are there any genes in Ensembl that contain somatic mutations associated with tumors in the eye?
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Somatic Variation (COSMIC 50) | Phenotype: COSMIC: tumor_site:eye | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| Phenotype description | ||
| Associated gene | ||
| Ensembl Gene ID |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Somatic Variation (COSMIC 50) | Phenotype: COSMIC: tumor_site:eye | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| Phenotype description | ||
| Associated gene | ||
| Ensembl Gene ID |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Somatic Variation (COSMIC 50) | Phenotype: COSMIC: tumor_site:eye | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| Phenotype description | ||
| Associated gene | ||
| Ensembl Gene ID |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens Somatic Variation (COSMIC 50) | Phenotype: COSMIC: tumor_site:eye | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| Phenotype description | ||
| Associated gene | ||
| Ensembl Gene ID |
The COSMIC project focuses on somatic mutations relating to human cancers. A somatic variation data set has been incorporated into the Ensembl Variation BioMart database to give users access to this data. A scientist can select from a list of COSMIC phenotypes from the GENERAL VARIATION FILTERS filter section, choose a selection of useful attributes from the Variation:SEQUENCE VARIATION and Variation:GENE ASSOCIATED INFORMATION attribute sections and export their results in a selection of file formats (Figure 3).
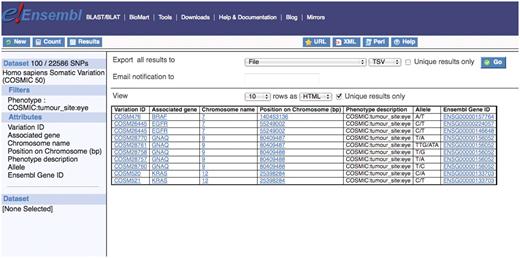
Shows that there are 100 single nucleotide polymorphisms in the human somatic variation data set associated with tumors in the eye and the list of Ensembl gene IDs containing these variations can be downloaded for further study or one can click on an entry in the Ensembl Gene ID column on the interface which links to the main Ensembl website.
Query #4: Find the HGNC symbols for a list of human variations.
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens variation (dbSNP 132;ENSEMBL) | Limit to variants with these IDs dbSNP rs IDs: rs348, rs362, rs364, rs565, rs645 | Variation ID |
| Chromosome name | ||
| Position on chromosome (bp) | ||
| Ensembl Gene ID | ||
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | HGNC ID | |
| HGNC symbol |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens variation (dbSNP 132;ENSEMBL) | Limit to variants with these IDs dbSNP rs IDs: rs348, rs362, rs364, rs565, rs645 | Variation ID |
| Chromosome name | ||
| Position on chromosome (bp) | ||
| Ensembl Gene ID | ||
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | HGNC ID | |
| HGNC symbol |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens variation (dbSNP 132;ENSEMBL) | Limit to variants with these IDs dbSNP rs IDs: rs348, rs362, rs364, rs565, rs645 | Variation ID |
| Chromosome name | ||
| Position on chromosome (bp) | ||
| Ensembl Gene ID | ||
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | HGNC ID | |
| HGNC symbol |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Variation 61: Homo sapiens variation (dbSNP 132;ENSEMBL) | Limit to variants with these IDs dbSNP rs IDs: rs348, rs362, rs364, rs565, rs645 | Variation ID |
| Chromosome name | ||
| Position on chromosome (bp) | ||
| Ensembl Gene ID | ||
| Ensembl Genes 61: Homo sapiens genes (GRCh37.p2) | HGNC ID | |
| HGNC symbol |
This query requires that the user selects filters and attributes from the human data set in the Variation BioMart database as well as selecting attributes from the human data set in the Ensembl Genes BioMart database. The linking of two data sets is a useful feature of the BioMart technology and allows for complex cross database queries to be constructed. In this query the user may have a list of dbSNP IDs and would like to obtain a list of Ensembl gene IDs and their corresponding HGNC IDs that contain these variations (Figure 4). The user must first upload their list of dbSNP IDs to the GENERAL VARIATION FILTERS section and then select the required attributes from the Variation:SEQUENCE VARIATION and Variation:GENE ASSOCIATED INFORMATION attribute sections. Then select the second data set [Homo sapiens genes (GRCh37.p2) from Ensembl Genes mart] from the left sidebar on the screen. Then select the HGNC ID and HGNC symbol from the features: EXTERNAL attribute section.
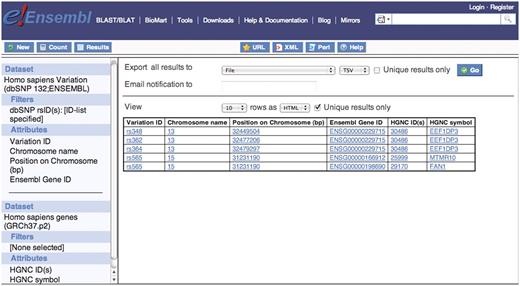
Five dbSNP rs IDs were used to filter the human variation data set and Ensembl gene IDs containing these five variations were selected in the attributes. Then linking to the second data set, human gene data set from Ensembl Genes database, the HGNC ID and symbol were selected in the attribute section to retrieve the corresponding gene names from HGNC. They are FAN1, MTMR10 and EEF1DP3.
Query #5: Find the genes from Escherichia coli strain K12 that are found within the region ‘360473–365601’ and discover whether there are any orthologs in the related strains E. coli O157:H7 EC4115 and E. coli DH10B.
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Bacteria Bacterial Mart (Release 8): Escherichia coli K12 genes | Gene start (bp): 360473 | Ensembl Gene ID |
| Gene end (bp): 365601 | Ensembl Transcript ID | |
| Associated Gene Name | ||
| Escherichia coli DH10B Ensembl Gene ID | ||
| Escherichia coli DH10B Chromosome Start (bp) | ||
| Escherichia coli DH10B Chromosome End (bp) | ||
| Escherichia coli O157:H7 EC4115 Ensembl Gene ID | ||
| Escherichia coli O157:H7 EC4115 Chromosome Start (bp) | ||
| Escherichia coli O157:H7 EC4115 Chromosome End (bp) |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Bacteria Bacterial Mart (Release 8): Escherichia coli K12 genes | Gene start (bp): 360473 | Ensembl Gene ID |
| Gene end (bp): 365601 | Ensembl Transcript ID | |
| Associated Gene Name | ||
| Escherichia coli DH10B Ensembl Gene ID | ||
| Escherichia coli DH10B Chromosome Start (bp) | ||
| Escherichia coli DH10B Chromosome End (bp) | ||
| Escherichia coli O157:H7 EC4115 Ensembl Gene ID | ||
| Escherichia coli O157:H7 EC4115 Chromosome Start (bp) | ||
| Escherichia coli O157:H7 EC4115 Chromosome End (bp) |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Bacteria Bacterial Mart (Release 8): Escherichia coli K12 genes | Gene start (bp): 360473 | Ensembl Gene ID |
| Gene end (bp): 365601 | Ensembl Transcript ID | |
| Associated Gene Name | ||
| Escherichia coli DH10B Ensembl Gene ID | ||
| Escherichia coli DH10B Chromosome Start (bp) | ||
| Escherichia coli DH10B Chromosome End (bp) | ||
| Escherichia coli O157:H7 EC4115 Ensembl Gene ID | ||
| Escherichia coli O157:H7 EC4115 Chromosome Start (bp) | ||
| Escherichia coli O157:H7 EC4115 Chromosome End (bp) |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Bacteria Bacterial Mart (Release 8): Escherichia coli K12 genes | Gene start (bp): 360473 | Ensembl Gene ID |
| Gene end (bp): 365601 | Ensembl Transcript ID | |
| Associated Gene Name | ||
| Escherichia coli DH10B Ensembl Gene ID | ||
| Escherichia coli DH10B Chromosome Start (bp) | ||
| Escherichia coli DH10B Chromosome End (bp) | ||
| Escherichia coli O157:H7 EC4115 Ensembl Gene ID | ||
| Escherichia coli O157:H7 EC4115 Chromosome Start (bp) | ||
| Escherichia coli O157:H7 EC4115 Chromosome End (bp) |
This query involves finding what E. coli genes lie in the given region and then discovering whether there are any orthologs in two related strains of E. coli. This is interesting as it may highlight bacterial genes that may have been acquired by some strains when compared to others and some genes may have been lost relative to other related strains (Figure 5). To do this query, add the gene start and end coordinates in the REGION filter section and then select the attributes from the Homologs:GENE and Homologs:ORTHOLOGS attribute sections.
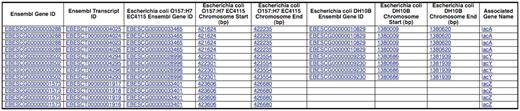
The genes in the filtered region were lacA, lacY and lacZ and we can see that there are no orthologs for the lacZ gene in the E. coli DH10B strain.
Query #6: The three-gene APL1 locus encodes essential components of the mosquito immune defense against malaria parasites. Find the variations within the APL1A, APL1B and APL1C genes as well as the strain name, strain genotype, allele and biotype.
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl MetazoaMetazoa Variation Mart (release 8): Anopheles gambiae variations (AgamP3) | Ensembl Gene IDs: AGAP007035 AGAP007036 AGAP007033 | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| dbSNP rsID | ||
| Strain Name | ||
| Strain Genotype | ||
| Ensembl Gene ID | ||
| Biotype |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl MetazoaMetazoa Variation Mart (release 8): Anopheles gambiae variations (AgamP3) | Ensembl Gene IDs: AGAP007035 AGAP007036 AGAP007033 | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| dbSNP rsID | ||
| Strain Name | ||
| Strain Genotype | ||
| Ensembl Gene ID | ||
| Biotype |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl MetazoaMetazoa Variation Mart (release 8): Anopheles gambiae variations (AgamP3) | Ensembl Gene IDs: AGAP007035 AGAP007036 AGAP007033 | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| dbSNP rsID | ||
| Strain Name | ||
| Strain Genotype | ||
| Ensembl Gene ID | ||
| Biotype |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl MetazoaMetazoa Variation Mart (release 8): Anopheles gambiae variations (AgamP3) | Ensembl Gene IDs: AGAP007035 AGAP007036 AGAP007033 | Variation ID |
| Chromosome name | ||
| Position on Chromosome (bp) | ||
| Allele | ||
| dbSNP rsID | ||
| Strain Name | ||
| Strain Genotype | ||
| Ensembl Gene ID | ||
| Biotype |
The Ensembl Metazoa Variation BioMart database consolidates single nucleotide polymorphisms from high-density, genome-wide mosquito SNP-genotyping array mapping and enables users to retrieve variations from the SNP-array identified through sequencing of two genetically diverged molecular forms of A. gambiae, Mopti (M) and Savanna (S) (23). This resource could help to analyze parasite susceptibility alleles from population subgroups. Query 6 shows how a user can obtain variation data for a particular gene or set of genes of interest (Figure 6). To do this query, the user must upload the gene IDs to the GENE ASSOCIATED VARIATION FILTERS section and then select the attributes of interest from the Variation: SEQUENCE VARIATION and Variation:GENE ASSOCIATED INFORMATION sections.
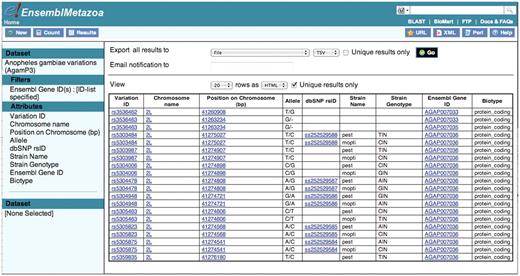
Having first retrieved the Ensembl gene IDs for the three APL1 genes, these are used to filter the A. gambiae data set. Fifty variations were retrieved that lie within the three genes of the APL1 locus.
Query #7: Find the coding sequence for all human genes on chromosome 22 along with the gene name and gene start and end.
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Gene 61: Homo sapiens genes (GRCh37.p2) | Chromosome 22 | Coding sequence |
| Ensembl Gene ID | ||
| Associated Gene Name | ||
| Associated Gene DB | ||
| Gene Start (bp) | ||
| Gene End (bp) |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Gene 61: Homo sapiens genes (GRCh37.p2) | Chromosome 22 | Coding sequence |
| Ensembl Gene ID | ||
| Associated Gene Name | ||
| Associated Gene DB | ||
| Gene Start (bp) | ||
| Gene End (bp) |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Gene 61: Homo sapiens genes (GRCh37.p2) | Chromosome 22 | Coding sequence |
| Ensembl Gene ID | ||
| Associated Gene Name | ||
| Associated Gene DB | ||
| Gene Start (bp) | ||
| Gene End (bp) |
| Database: Data sets . | Filters . | Attributes . |
|---|---|---|
| Ensembl Gene 61: Homo sapiens genes (GRCh37.p2) | Chromosome 22 | Coding sequence |
| Ensembl Gene ID | ||
| Associated Gene Name | ||
| Associated Gene DB | ||
| Gene Start (bp) | ||
| Gene End (bp) |
The BioMart technology allows for the download of sequence information in a usable format. This is a powerful feature that allows users to retrieve flanking sequence, exon sequence, 3′ and 5′-UTR, cDNA sequence, coding sequence and protein sequence. Query 7 illustrates how to retrieve coding sequences for all genes on chromosome 22 as well as obtaining information about the gene name and the location of the gene start and end (Figure 7). To do this query, select the chromosome from the REGION filter section and the attributes of interest from the Sequences:SEQUENCES and Sequence:Header Information attribute sections.
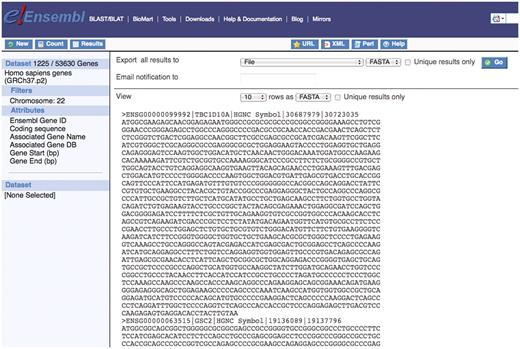
The ability to retrieve sequence information for genes of interest is a powerful feature of the BioMart tool. Here a user can download the coding sequence for all genes on chromosome 22 as well as additional information about each gene and this can be exported in a useful format.
Discussion and future directions
The BioMart interface and querying platform provides the Ensembl and Ensembl Genomes projects with the necessary tools to design BioMart databases from the various source databases produced by the project. The BioMart databases and accompanying interface provides users with a fast and flexible means of querying the customized sets of biological data using a wide range of querying methods. The BioMart software also allows federation to other databases of scientific interest so that cross querying can be accomplished. It also allows the Ensembl and Ensembl Genomes databases to be incorporated into other portals with ease such as www.biomart.org.
As scientific activity evolves and in an effort to provide the most useful resources for our users, both the Ensembl and Ensembl Genomes projects will incorporate data from additional species and additionally handle new types of data, which will be included in the project BioMarts. In the future, we plan to move both projects to the new BioMart 0.8 code (24) and incorporate the new interface into the main Ensembl website.
Funding
The Wellcome Trust provide majority funding for the Ensembl project (grant number WT062023) with additional support from the European Commission under SLING, grant agreement number 226073 (Integrating Activity) within Research Infrastructures of the FP7 Capacities Specific Programme; the UK Biotechnology and Biological Sciences Research Council (grant numbers BB/F019793/1, BB/I001077/1); the Bill and Melinda Gates Foundation; and the European Molecular Biological Laboratory. Funding for open access charge: The Wellcome Trust.
Conflict of interest. None declared.
Acknowledgements
The authors thank all the users of the Ensembl and Ensembl Genomes projects especially those who have provided us with feedback about the Ensembl BioMarts. The authors would also like to thank the members of the BioMart team at the Ontario Institute for Cancer Research (OICR), especially Dr Arek Kasprzyk, for providing sustained technical support and assistance over the years.
References
Author notes
Present address: Jorge Zamora, Structural Computational Biology Group, Spanish National Cancer Research Centre, C/ Melchor Fernández Almagro, 3, 28029, Madrid, Spain



