-
PDF
- Split View
-
Views
-
Cite
Cite
Darren J. Oakley, Vivek Iyer, William C. Skarnes, Damian Smedley, BioMart as an integration solution for the International Knockout Mouse Consortium, Database, Volume 2011, 2011, bar028, https://doi.org/10.1093/database/bar028
Close - Share Icon Share
Abstract
In this article, we describe the use of the BioMart data management system to provide integrated access to International Knockout Mouse Consortium (IKMC) data and other related mouse resources. The IKMC is currently mutating all mouse protein-coding genes in embryonic stem (ES) cells using gene targeting and gene trapping approaches. The BioMart portal allows researchers to identify and obtain IKMC knockout vectors, ES cells and mice for genes of interest. Gene annotation, expression, phenotype and disease data is also integrated from external BioMarts, allowing selection of IKMC products by a wide variety of criteria. These products are invaluable for researchers involved in the elucidation of gene function and the role of individual genes in human disease. Here, we describe these datasets in more detail and illustrate the functionality of the portal using several examples.
Database URL: http://www.knockoutmouse.org/mart
Project description
The International Knockout Mouse Consortium [IKMC; (1)] was formed shortly after the completion of the mouse genome sequence with the aim of mutating all mouse protein-coding genes in ES cells. These ES cells can be used to produce knockout mice for phenotyping. The mouse is the premier model organism due to its accessibility to genetic and phenotype analysis and similarity to humans. The IKMC therefore provides an invaluable, time and cost effective resource for scientists studying disease mechanisms as well as mammalian gene function.
The IKMC consists of KOMP [KnockOut Mouse Project; (2)] funded by the National Institutes of Health (NIH, USA); EUCOMM [(3); EUropean Conditional Mouse Mutagenesis Program] funded by the European Commission (EC), NorCOMM (North American Conditional Mouse Mutagenesis Project) funded by Genome Canada and the Texas Institute of Genomic Medicine [TIGM; (4)]. All material produced from KOMP is distributed by the KOMP repository (www.komp.org), NorCOMM material by the Canadian Mouse Mutant Repository at the Toronto Centre for Phenogenomics (www.phenogenomics.ca), EUCOMM vectors and ES cells by the European Mouse Mutant Cell Repository (www.eummcr.org) and mice produced from EUCOMM resources are supplied from the European Mouse Mutant Archive [EMMA; (5)] while TIGM distributes its own ES cells and mice.
The IKMC web portal (www.knockoutmouse.org) has been created to provide access to all data and resources as well as to coordinate and track the efforts of the various IKMC projects and prioritize the genes to target (6). This portal is jointly maintained by the KOMP Data Coordination Center (KOMP-DCC) funded by the NIH and the I-DCC (International-Data Coordination Center) funded by the EC. A centralized database currently provides data access, but to facilitate long-term maintenance and integrate additional biological information we have adopted BioMart technology [www.biomart.org; (7,8); J. Zhang et al., submitted for publication]. BioMart is generic software that can easily be deployed on existing data resources distributed in different geographical locations to allow integrated querying via web services. BioMart therefore provides a distributed and federated alternative to a central data warehouse and has been deployed on some 40+ publicly accessible BioMart databases around the world. Our BioMart portal is accessible through www.knockoutmouse.org/biomart/martview.
Data content
The BioMart portal at IKMC allows integrated querying of datasets created especially for the IKMC as well as external BioMarts produced by other mouse resources.
To date, the IKMC has produced mutant ES cell lines for 16 031 genes out of the predicted 25 000 protein coding genes in C57BL/6N ES cells. At least 947 of these have been used to produce knockout mice that can be ordered from the various IKMC repositories. The pipelines creating targeted mutations proceed from gene annotation to vector design and vector construction, targeting in ES cells and microinjection or aggregation to produce mutant mice. Each stage is associated with quality control steps. Additionally, each of the projects uses different and complementary targeting strategies (http://www.knockoutmouse.org/about/targeting-strategies).
The IKMC projects/alleles dataset provides data on each mutation attempt including the mutated gene, chromosomal location and synonyms/external resource IDs for the gene, which project is carrying out the mutation and the status in the pipeline and whether vector, ES cells and mice are available for researchers to order. In addition, the number of gene traps, targeted and other types of mutants for the same gene in non-IKMC resources is also noted. The content of this dataset is derived wholly from the IDCC master gene list and targeting project data maintained by MGI (6).
The targeted products dataset catalogues the targeted mutations (vectors and ES cells) produced by IKMC pipelines. The set includes vector details such as the exon removed by the targeting, genomic positions of mutagenic insertions and links to annotated mutant sequence, as well as detailed, publication quality diagrams of vectors and alleles. Quality control data on the ES cell clones arising from the production center, distribution center and users of the clones are also recorded along with the officially assigned ID for the mutant allele.
The IKMC mouse production dataset tracks the next stage of the process where the mutant ES cells are microinjected or aggregated to eventually produce a mutant mouse line. This dataset captures the breeding status of the colony, the ES cell clone and allele details along with the inbred mouse strains used for the microinjection or aggregation, test crossing and back crossing. In addition, the results of quality control steps such as Southern blotting to confirm the correct mutation in the final line are noted.
All the datasets available from the IKMC BioMart portal, including external datasets, are summarized in Table 1.
| Mart . | Dataset . | Description of data content . |
|---|---|---|
| IKMC genes and products | IKMC projects/alleles | IKMC project and status for each gene as well as availability of vectors, ES cells and mice |
| IKMC genes and products | IKMC targeted products | Details on IKMC vector design and tracking of production from vector to ES cells |
| IKMC genes and products | IKMC mouse production | Tracking of IKMC mouse production |
| UniTrap | UniTrap | Gene trap data from UniTrap (9) |
| IKMC genes and products | OMIM | Online Mendelian Inheritance in Man (OMIM) disease and gene data including mouse orthologues produced by merging the OMIM gene map with a list of mouse orthologues downloaded from the Mouse Genomics Informatics group at the Jackson Laboratory (10,11) |
| WTSI Mouse Genetics Project (Sanger UK) | MGP phenotyping | High-throughput phenotype and expression data from the Wellcome Trust Sanger Institute Mouse Genetics Project (MGP) |
| European Mouse Mutant Archive | EMMA strains | EMMA (main repository for European mutant mice including those produced from EUCOMM ES cell resources) data from the BioMart at www.emmanet.org/biomart/martview (5) |
| The CREATE consortium | Cre lines | Virtual repository of Cre-recombinase expressing lines required for conditional knocking out of IKMC genes in a spatial and temporally controlled manner (www.creline.org/biomart/martview) |
| Eurexpress | Eurexpress | In situ embryonic expression data from the BioMart at biomart.eurexpress.org (12) |
| Europhenome | Europhenome | High-throughput phenotype data of mice produced from EUCOMM ES cell resources from the BioMart at www.europhenome.org/biomart/martview (13) |
| Ensembl Gene (Sanger UK) | Ensembl Mus musculus genes | Mouse genes with annotated external references, protein domains, orthologues, variation and genomic data from the BioMart of the Ensembl project at www.ensembl.org/biomart/martview (14) |
| Vega (Sanger UK) | Vega Mus musculus genes | Manually curated mouse genes from the BioMart at www.ensembl.org/biomart/martview (14) |
| MGI (Jackson Laboratory US) | Features | Curated genes and alleles from the BioMart of the MGI group at biomart.informatics.jax.org (11) |
| Mart . | Dataset . | Description of data content . |
|---|---|---|
| IKMC genes and products | IKMC projects/alleles | IKMC project and status for each gene as well as availability of vectors, ES cells and mice |
| IKMC genes and products | IKMC targeted products | Details on IKMC vector design and tracking of production from vector to ES cells |
| IKMC genes and products | IKMC mouse production | Tracking of IKMC mouse production |
| UniTrap | UniTrap | Gene trap data from UniTrap (9) |
| IKMC genes and products | OMIM | Online Mendelian Inheritance in Man (OMIM) disease and gene data including mouse orthologues produced by merging the OMIM gene map with a list of mouse orthologues downloaded from the Mouse Genomics Informatics group at the Jackson Laboratory (10,11) |
| WTSI Mouse Genetics Project (Sanger UK) | MGP phenotyping | High-throughput phenotype and expression data from the Wellcome Trust Sanger Institute Mouse Genetics Project (MGP) |
| European Mouse Mutant Archive | EMMA strains | EMMA (main repository for European mutant mice including those produced from EUCOMM ES cell resources) data from the BioMart at www.emmanet.org/biomart/martview (5) |
| The CREATE consortium | Cre lines | Virtual repository of Cre-recombinase expressing lines required for conditional knocking out of IKMC genes in a spatial and temporally controlled manner (www.creline.org/biomart/martview) |
| Eurexpress | Eurexpress | In situ embryonic expression data from the BioMart at biomart.eurexpress.org (12) |
| Europhenome | Europhenome | High-throughput phenotype data of mice produced from EUCOMM ES cell resources from the BioMart at www.europhenome.org/biomart/martview (13) |
| Ensembl Gene (Sanger UK) | Ensembl Mus musculus genes | Mouse genes with annotated external references, protein domains, orthologues, variation and genomic data from the BioMart of the Ensembl project at www.ensembl.org/biomart/martview (14) |
| Vega (Sanger UK) | Vega Mus musculus genes | Manually curated mouse genes from the BioMart at www.ensembl.org/biomart/martview (14) |
| MGI (Jackson Laboratory US) | Features | Curated genes and alleles from the BioMart of the MGI group at biomart.informatics.jax.org (11) |
| Mart . | Dataset . | Description of data content . |
|---|---|---|
| IKMC genes and products | IKMC projects/alleles | IKMC project and status for each gene as well as availability of vectors, ES cells and mice |
| IKMC genes and products | IKMC targeted products | Details on IKMC vector design and tracking of production from vector to ES cells |
| IKMC genes and products | IKMC mouse production | Tracking of IKMC mouse production |
| UniTrap | UniTrap | Gene trap data from UniTrap (9) |
| IKMC genes and products | OMIM | Online Mendelian Inheritance in Man (OMIM) disease and gene data including mouse orthologues produced by merging the OMIM gene map with a list of mouse orthologues downloaded from the Mouse Genomics Informatics group at the Jackson Laboratory (10,11) |
| WTSI Mouse Genetics Project (Sanger UK) | MGP phenotyping | High-throughput phenotype and expression data from the Wellcome Trust Sanger Institute Mouse Genetics Project (MGP) |
| European Mouse Mutant Archive | EMMA strains | EMMA (main repository for European mutant mice including those produced from EUCOMM ES cell resources) data from the BioMart at www.emmanet.org/biomart/martview (5) |
| The CREATE consortium | Cre lines | Virtual repository of Cre-recombinase expressing lines required for conditional knocking out of IKMC genes in a spatial and temporally controlled manner (www.creline.org/biomart/martview) |
| Eurexpress | Eurexpress | In situ embryonic expression data from the BioMart at biomart.eurexpress.org (12) |
| Europhenome | Europhenome | High-throughput phenotype data of mice produced from EUCOMM ES cell resources from the BioMart at www.europhenome.org/biomart/martview (13) |
| Ensembl Gene (Sanger UK) | Ensembl Mus musculus genes | Mouse genes with annotated external references, protein domains, orthologues, variation and genomic data from the BioMart of the Ensembl project at www.ensembl.org/biomart/martview (14) |
| Vega (Sanger UK) | Vega Mus musculus genes | Manually curated mouse genes from the BioMart at www.ensembl.org/biomart/martview (14) |
| MGI (Jackson Laboratory US) | Features | Curated genes and alleles from the BioMart of the MGI group at biomart.informatics.jax.org (11) |
| Mart . | Dataset . | Description of data content . |
|---|---|---|
| IKMC genes and products | IKMC projects/alleles | IKMC project and status for each gene as well as availability of vectors, ES cells and mice |
| IKMC genes and products | IKMC targeted products | Details on IKMC vector design and tracking of production from vector to ES cells |
| IKMC genes and products | IKMC mouse production | Tracking of IKMC mouse production |
| UniTrap | UniTrap | Gene trap data from UniTrap (9) |
| IKMC genes and products | OMIM | Online Mendelian Inheritance in Man (OMIM) disease and gene data including mouse orthologues produced by merging the OMIM gene map with a list of mouse orthologues downloaded from the Mouse Genomics Informatics group at the Jackson Laboratory (10,11) |
| WTSI Mouse Genetics Project (Sanger UK) | MGP phenotyping | High-throughput phenotype and expression data from the Wellcome Trust Sanger Institute Mouse Genetics Project (MGP) |
| European Mouse Mutant Archive | EMMA strains | EMMA (main repository for European mutant mice including those produced from EUCOMM ES cell resources) data from the BioMart at www.emmanet.org/biomart/martview (5) |
| The CREATE consortium | Cre lines | Virtual repository of Cre-recombinase expressing lines required for conditional knocking out of IKMC genes in a spatial and temporally controlled manner (www.creline.org/biomart/martview) |
| Eurexpress | Eurexpress | In situ embryonic expression data from the BioMart at biomart.eurexpress.org (12) |
| Europhenome | Europhenome | High-throughput phenotype data of mice produced from EUCOMM ES cell resources from the BioMart at www.europhenome.org/biomart/martview (13) |
| Ensembl Gene (Sanger UK) | Ensembl Mus musculus genes | Mouse genes with annotated external references, protein domains, orthologues, variation and genomic data from the BioMart of the Ensembl project at www.ensembl.org/biomart/martview (14) |
| Vega (Sanger UK) | Vega Mus musculus genes | Manually curated mouse genes from the BioMart at www.ensembl.org/biomart/martview (14) |
| MGI (Jackson Laboratory US) | Features | Curated genes and alleles from the BioMart of the MGI group at biomart.informatics.jax.org (11) |
Query examples
A description of BioMart aimed at biologist users has recently been published (7) so we will not describe all of BioMart’s querying options here. In summary, every BioMart query involves choosing one or more ‘Datasets’ by clicking on the datasets tab in the left hand panel and choosing from the drop-down followed by choosing ‘Filters’ to limit the records that are returned and ‘Attributes’ that correspond to the columns in the final results table. Filters and attributes are set by clicking on the tabs in the left hand panel and making selections in the right hand panel. A summary of the query is presented in the left hand panel and once complete, a preview of the results is generated by clicking on the ‘Results’ button. From this preview, the output format can be changed and a full download of the results performed.
To demonstrate the utility of the IKMC BioMart Portal we present several biologically relevant queries that can be performed using the datasets at www.knockoutmouse.org/biomart/martview.
Query 1: ‘Find all IKMC resources for genes encoding transcription factors on chromosome 1 between 180-190 Mbp’.
| Dataset . | Filters . | Attributes . |
|---|---|---|
| Mus musculus genes (NCBIM37) | GO term name: transcription regulator activity | GO term name |
| Chromosome: 1 | GO term accession | |
| Gene start (bp): 180000000 | Chromosome name | |
| Gene end (bp): 190000000 | Gene start | |
| IKMC projects/Alleles | ES cell available : yes | Marker symbol |
| IKMC project/pipeline : KOMP-CSD, KOMP-Regeneron, EUCOMM,NorCOMM | MGI accession ID | |
| IKMC project | ||
| IKMC project ID | ||
| Status | ||
| Mouse available | ||
| ES cell available | ||
| Vector available |
| Dataset . | Filters . | Attributes . |
|---|---|---|
| Mus musculus genes (NCBIM37) | GO term name: transcription regulator activity | GO term name |
| Chromosome: 1 | GO term accession | |
| Gene start (bp): 180000000 | Chromosome name | |
| Gene end (bp): 190000000 | Gene start | |
| IKMC projects/Alleles | ES cell available : yes | Marker symbol |
| IKMC project/pipeline : KOMP-CSD, KOMP-Regeneron, EUCOMM,NorCOMM | MGI accession ID | |
| IKMC project | ||
| IKMC project ID | ||
| Status | ||
| Mouse available | ||
| ES cell available | ||
| Vector available |
A researcher may have narrowed down their search for candidates gene(s) involved in a disease of interest and identified the orthologous mouse region as between 180 and 190Mbp of chromosome 1. In addition, they may expect the gene to be a transcription factor and to test the potential candidates they have decided to produce mouse knockouts and phenotype them to test whether the mice share features of the disease of interest.
Query 1 identifies genes encoding transcription factors in this region and the IKMC project that is producing knockouts of these genes, the status of this pipeline and whether vector, ES cells or a full mouse knockout is available to order. Query 1 is set up at the interface at www.knockoutmouse.org/mart by first of all selecting the Mus musculus genes (NCBI37) Dataset from Ensembl genes. The second IKMC Project/Alleles Dataset required for this query is chosen by clicking on the bottom of the two Dataset tabs in the left hand panel (see Figure 1). Filters and Attributes are set by clicking on the Filter and Attribute tabs (again in the left hand panel shown in Figure 1) and selecting the relevant options that appear in the main right hand panel. Clicking the Results button in the top menu bar will reveal a preview of the results in the main right hand panel (Figure 1). In this case, a mouse knockout is already available for the researcher.
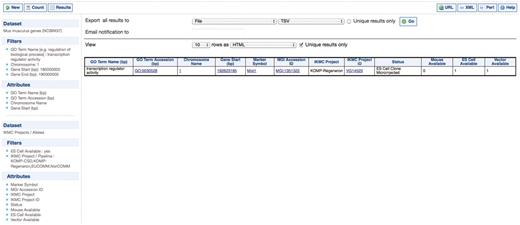
IKMC resources for genes encoding transcription factors on Chromosome 1 between 180 and 190 Mbp. This query involves the Mus musculus genes dataset from Ensembl and the IKMC projects/alleles dataset.
Query 2: ‘Find all IKMC resources for genes expressed in heart’.
| Dataset . | Filters . | Attributes . |
|---|---|---|
| Eurexpress Biomart | EMAP Term : heart | Pattern |
| EMAP Term | ||
| Strength | ||
| Assay ID | ||
| IKMC projects/alleles | ES cell available : yes | Marker symbol |
| IKMC project/pipeline : KOMP-CSD, KOMP-Regeneron, EUCOMM,NorCOMM | MGI accession ID | |
| IKMC project | ||
| IKMC project ID | ||
| Status | ||
| Mouse available | ||
| ES cell available | ||
| Vector available |
| Dataset . | Filters . | Attributes . |
|---|---|---|
| Eurexpress Biomart | EMAP Term : heart | Pattern |
| EMAP Term | ||
| Strength | ||
| Assay ID | ||
| IKMC projects/alleles | ES cell available : yes | Marker symbol |
| IKMC project/pipeline : KOMP-CSD, KOMP-Regeneron, EUCOMM,NorCOMM | MGI accession ID | |
| IKMC project | ||
| IKMC project ID | ||
| Status | ||
| Mouse available | ||
| ES cell available | ||
| Vector available |
Query 2 identifies genes expressed in the heart or sub-structures and which IKMC project is producing knockouts of these genes, the status of this pipeline and whether vector, ES cells or a full mouse knockout is available to order (Figure 2). Using this query, a scientist interested in the role of these genes in heart disease or heart development could identify IKMC resources to facilitate his/her research.
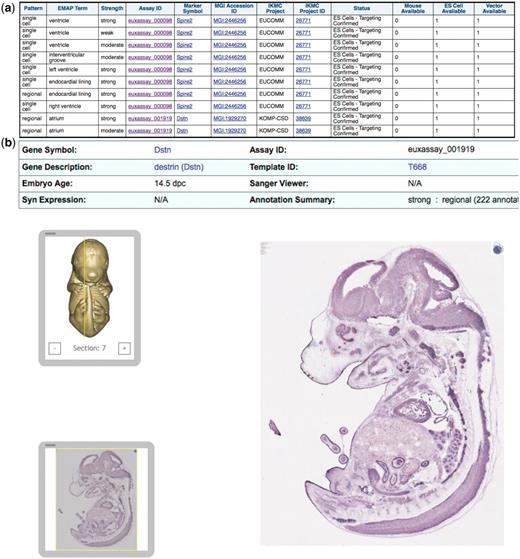
IKMC resources for genes expressed in embryonic heart or sub-structures according to the Eurexpress in situ expression database: (a) results of a query involving the in situ expression dataset from Eurexpress and the IKMC projects/alleles dataset, (b) further detail of the expression patterns at the main Eurexpress website after linking from one of the Assay ID results above.
Query 3: ‘Find all IKMC mice available from the EMMA Repository with information on the vector used to make the mutation’.
| Dataset . | Filters . | Attributes . |
|---|---|---|
| IKMC mouse production | Available from EMMA? : yes | Marker symbol |
| Microinjection status : genotype confirmed | Microinjection centre | |
| Sponsor : EUCOMM | Sponsor | |
| IKMC targeted products | Cassette | |
| Allele symbol superscript | ||
| ES cell clone | ||
| Targeting vector | ||
| Parental cell line | ||
| Mutation type | ||
| Mutation subtype |
| Dataset . | Filters . | Attributes . |
|---|---|---|
| IKMC mouse production | Available from EMMA? : yes | Marker symbol |
| Microinjection status : genotype confirmed | Microinjection centre | |
| Sponsor : EUCOMM | Sponsor | |
| IKMC targeted products | Cassette | |
| Allele symbol superscript | ||
| ES cell clone | ||
| Targeting vector | ||
| Parental cell line | ||
| Mutation type | ||
| Mutation subtype |
This query identifies mice available from the EMMA repository that have been generated from EUCOMM ES cell resources along with the mouse centre that produced the mouse resource and details on the vector used to generate this line. This extra detail includes the cassette used in the vector, which parental ES Cell line was used, mutation type and subtype as well as links to more detailed pages on the ES cell clone and targeting vector (Figure 3).
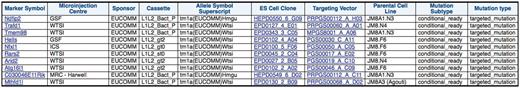
IKMC mice available from the EMMA Repository and details of the vector used to make the knockout. This query involves the IKMC mouse production and IKMC targeted products dataset.
Query 4: ‘Show me all the distributed EMMA lines have passed Southern blot quality control at a distribution center’.
| Dataset . | Filters . | Attributes . |
|---|---|---|
| EMMA strains | EMMA ID | |
| Gene symbol | ||
| Allele symbol | ||
| IKMC mouse production | Southern blot: pass | Sponsor |
| ES cell clone | ||
| Microinjection centre | ||
| Microinjection status |
| Dataset . | Filters . | Attributes . |
|---|---|---|
| EMMA strains | EMMA ID | |
| Gene symbol | ||
| Allele symbol | ||
| IKMC mouse production | Southern blot: pass | Sponsor |
| ES cell clone | ||
| Microinjection centre | ||
| Microinjection status |
Certain quality control checks are performed after mouse production. This query identifies EMMA lines that have passed a Southern blot check (Figure 4).
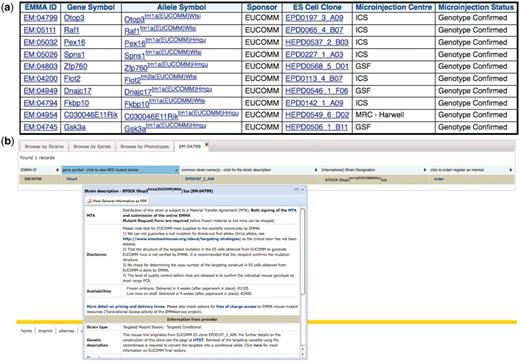
Distributed EMMA lines that have passed Southern blot quality control at a distribution center: (a) results of a query involving the dataset from the mouse distribution dataset at the European Mouse Mutant Archive (EMMA) and the IKMC mouse production dataset, (b) further detail of one of the mouse lines at the main EMMA website after linking from one of the above results.
Query 5: ‘Is there any existing phenotype data for other mouse knockouts of the same gene for mouse lines produced from EUCOMM ES resources’.
| Dataset . | Filters . | Attributes . |
|---|---|---|
| MGI features | Allele symbol | |
| Phenotype term | ||
| IKMC mouse production | Sponsor : EUCOMM | Marker symbol |
| Available from EMMA? : yes | Allele name | |
| Microinjection status : Genotype confirmed |
| Dataset . | Filters . | Attributes . |
|---|---|---|
| MGI features | Allele symbol | |
| Phenotype term | ||
| IKMC mouse production | Sponsor : EUCOMM | Marker symbol |
| Available from EMMA? : yes | Allele name | |
| Microinjection status : Genotype confirmed |
Before ordering a mouse line produced from IKMC resources, a researcher may be interested if other knockouts of the same gene have previously been generated and phenotyped. The MGI group curates all publications on mouse models including the phenotype descriptions using the Mammalian Phenotype ontology. Using the above query, a scientist can retrieve this data for each of the IKMC resources (Figure 5) and evaluate whether the model may be useful for their research bearing in mind that the phenotype can vary with the particular mutant allele and genetic background of the line.
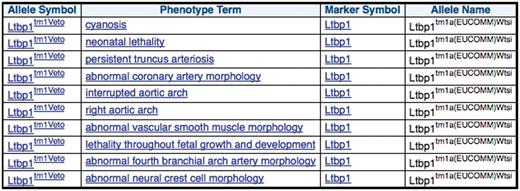
MGI curated phenotype data for other mutant alleles of the same gene for each of the mouse lines produced from EUCOMM ES cell resources.
Discussion and future directions
In this article, we have demonstrated how BioMart is a useful way of exposing the data being generated by the IKMC. In particular, it allows researchers to query on a wide range of criteria and identify physical resources such as vectors, ES cells and mouse lines to order from the repositories associated with the IKMC. BioMart also provides integrated querying of gene annotation, expression, phenotype and disease data from external databases. The example queries in this manuscript show how Ensembl, MGI and Eurexpress can be queried alongside IKMC data to potentially identify resources for a researcher to order.
In addition, the use of BioMart as a software solution has been hugely beneficial to us. The alternative approach of generating and maintaining an up to date, centralized database containing all the data presented in the IKMC BioMart portal would be a huge technical and social challenge. IKMC consists of four, separately funded projects and, for EUCOMM alone; there are five separate centers generating the knockout mice and six centers distributing them. The high-throughput nature of the strategy requires automation of data entry and exchange and each of these centers has its own informatics tracking systems. Just solving the technical challenges such as data exchange formats and tracking all the data exchange is a huge challenge. Centrally warehousing the external data we present at the BioMart portal would also raise its own social issues such as appropriate attribution and coordination of data releases.
In the immediate future, we hope to expand the range of querying possible from the IKMC BioMart portal by including two new datasets currently being developed by members of the I-DCC. The first of these will expose data from the International Mouse Strain Resource [IMSR; (15)]. The IMSR catalogues mutant mice held in public repositories around the world and therefore having this dataset will allow us to present alternative sources for knockout mice, in particular where IKMC material does not yet exist. The second will integrate expression data from the Gene Expression Database [GXD; (16)]. GXD collects mouse expression data from a wide range of life stages and experimental techniques.
Looking further ahead, one exciting new source of data will be that emerging from the International Mouse Phenotyping Consortium (IMPC). The IMPC plans to use IKMC resources to generate knockout mice for every single protein-coding gene and then phenotype them using standardized, high-throughput pipelines. The analysed and annotated data are likely to be very similar to that already in the Europhenome and Wellcome Trust Sanger Institute Mouse Genetics Project datasets, but cover all genes rather than the few hundred currently presented. Having a BioMart of this phenotype data will allow integrated querying with all the other mouse resources already at the IKMC portal, allowing researchers to identify and order useful IMPC mice for further characterization in their individual laboratories.
To provide a larger range of query interfaces we are planning to update our site to the new BioMart 0.8 software. In addition, we are developing our own query interface that combines BioMart web service querying with Apache Solr indexing. This interface will allow simple, Google-like searching across all the data held in the various BioMarts using terms such as gene names or anatomical or phenotype terms. This prototype will be released on the IKMC web portal later in this year.
Funding
International-Data Coordination Centre for the IKMC (European Commission project 223592). Funding for open access charge: European Commission project 223592.
Conflict of interest. None declared.
Acknowledgements
We are most grateful to all the members of the IKMC programs for making the data available for creation of the BioMart datasets. In particular, we thank Jeremy C. Mason, Kevin Stone, James A. Kadin, Janan T. Eppig and Martin Ringwald from the Jackson Laboratory, Bar Harbor, USA. In addition, we thank the external projects who created the additional BioMart datasets used in the integrated querying examples described in this article including Richard Baldock and Bernard Haggarty from the Eurexpress project, Andrew Blake and Ann-Marie Mallon from Europhenome, Matthew Hall from the MGI group and Rhoda Kinsella from Ensembl.



