-
PDF
- Split View
-
Views
-
Cite
Cite
Rebecca Shepherd, Simon A. Forbes, David Beare, S. Bamford, Charlotte G. Cole, Sari Ward, Nidhi Bindal, Prasad Gunasekaran, Mingming Jia, Chai Yin Kok, Kenric Leung, Andrew Menzies, Adam P. Butler, Jon W. Teague, Peter J. Campbell, Michael R. Stratton, P. Andrew Futreal, Data mining using the Catalogue of Somatic Mutations in Cancer BioMart, Database, Volume 2011, 2011, bar018, https://doi.org/10.1093/database/bar018
Close - Share Icon Share
Abstract
Catalogue of Somatic Mutations in Cancer (COSMIC) (http://www.sanger.ac.uk/cosmic) is a publicly available resource providing information on somatic mutations implicated in human cancer. Release v51 (January 2011) includes data from just over 19 000 genes, 161 787 coding mutations and 5573 gene fusions, described in more than 577 000 tumour samples. COSMICMart (COSMIC BioMart) provides a flexible way to mine these data and combine somatic mutations with other biological relevant data sets. This article describes the data available in COSMIC along with examples of how to successfully mine and integrate data sets using COSMICMart.
Database URL: http://www.sanger.ac.uk/genetics/CGP/cosmic/biomart/martview/
Project description
COSMIC is a repository for somatic mutations and associated phenotype/clinical information, combining data from a number of sources. Primarily, data are manually curated from the scientific literature for genes selected from the Cancer Gene Census (http://www.sanger.ac.uk/genetics/CGP/Census/), a listing of genes that are known to be mutated in human cancer. The manual curation initially focused on known cancer genes that had a high proportion of coding point mutations. In recent years, this has been extended to include cancer genes that are also mutated by gene fusion events. With the advent of next-generation sequencing, COSMIC has been adapted to hold complete catalogues of somatic mutations for individual tumour samples.
The COSMIC website has been developed to overview the underlying database in a user-friendly manner. The website can be navigated by gene, cancer sample or tissue/histology type and has a series of graphical and tabular displays to summarize the content of the database.
However, the integration of biological data sets is still a major IT challenge. Although, the data in COSMIC can be viewed and downloaded in a number of useful ways, it is still challenging for users to generate their own custom queries and also to link to related resources. In order to facilitate the integration of the COSMIC data set, we have used the BioMart data mining software. BioMart uses a federated data model to allow integration of biological data from diverse databases (1). There is now a significant number of data resources that have set up their own BioMarts including Ensembl, UniProt and HGNC. The BioMart software also has interfaces that allow users to easily generate custom queries to obtain subsets of data from one or more BioMart data resources. We have successfully set up an instance of a BioMart, COSMICMart, which holds a summary of the somatic mutations and associated phenotype data from the COSMIC database.
Data content
COSMIC version 51 (January 2011) contains a full curation of the scientific literature for 91 cancer genes, mostly point-mutated, as well as 53 curated fusion gene pairs, with over 11 000 papers having been assessed. Weekly literature searches are carried out for each cancer gene and each abstract/article is manually evaluated. Checks are made to ensure a paper has not been curated already, whether the paper is a review or is reporting original data, if there are any inconsistencies in the data, and whether all the required information for full curation is present, e.g. information on the numbers of samples screened and sufficient mutation detail. Papers which pass the initial scan are submitted for detailed manual curation, while the remaining papers are ‘listed’ on the COSMIC website. Information is manually recorded on the actual mutation change, the detailed phenotype of the cancer samples and the original publication/study.
Mutation changes are checked for accuracy and recorded at the genome, transcript (to one reference transcript) and protein level using the mutation syntax standards developed by the Human Genome Variation Society (2). Negative results (sample screened does not have a mutation for a particular gene for a particular publication) are also recorded so prevalence statistics can be estimated for each gene. As part of the curation process, cancer sample tissue/histology descriptions from the original publication are recorded and then re-classified to a COSMIC standard tissue/histology ontology (See classification section of the COSMIC additional information page, http://www.sanger.ac.uk/genetics/CGP/cosmic/add_info/). The standardization of the main data types in COSMIC means the utility of the data is significantly enhanced and the data are easier to browse and incorporate into external data sets. When available, clinical and exposure data and therapeutic responses are also recorded. The full functionality of COSMIC has previously been described (3, 4).
The database coverage has recently been enhanced by the integration of somatic mutations from external data resources. A collaboration with the database curators at International Agency for Research on Cancer (IARC) has allowed the majority of mutations from IARC TP53 database R14 to be integrated into COSMIC (5). Somatic mutations from large-scale systematic cancer screens are curated in COSMIC to include studies on glioblastoma multiforme, breast, colorectal and lung cancer (6–9). Data are also directly curated from the data portals of large-scale projects to include the Cancer Genome Project (CGP) at the Sanger Institute and more recently validated somatic mutations from The Cancer Genome Atlas (TCGA) (6) and International Cancer Genome Consortium (ICGC) (10). COSMIC has been enhanced to accommodate somatic mutations from whole genome and exome screens. This includes all somatic and non-coding mutations, structural rearrangements and gene fusions. Currently, COSMIC has annotated 51 genomes and 332 exomes from a range of cancers to include lung, malignant melanoma, renal, AML, pancreatic and ovarian cancer. The current contents of the database (v51, January 2011) are displayed in Table 1.
| Curated data type . | Curated data count . |
|---|---|
| Experiments | 2 946 792 |
| Tumours | 577 304 |
| Mutations | 167 193 |
| References | 11 062 |
| Genes | 19 000 |
| Fusions | 5573 |
| Structural variants | 2729 |
| Whole-cancer genomes | 51 |
| Whole-cancer exomes | 332 |
| Curated data type . | Curated data count . |
|---|---|
| Experiments | 2 946 792 |
| Tumours | 577 304 |
| Mutations | 167 193 |
| References | 11 062 |
| Genes | 19 000 |
| Fusions | 5573 |
| Structural variants | 2729 |
| Whole-cancer genomes | 51 |
| Whole-cancer exomes | 332 |
| Curated data type . | Curated data count . |
|---|---|
| Experiments | 2 946 792 |
| Tumours | 577 304 |
| Mutations | 167 193 |
| References | 11 062 |
| Genes | 19 000 |
| Fusions | 5573 |
| Structural variants | 2729 |
| Whole-cancer genomes | 51 |
| Whole-cancer exomes | 332 |
| Curated data type . | Curated data count . |
|---|---|
| Experiments | 2 946 792 |
| Tumours | 577 304 |
| Mutations | 167 193 |
| References | 11 062 |
| Genes | 19 000 |
| Fusions | 5573 |
| Structural variants | 2729 |
| Whole-cancer genomes | 51 |
| Whole-cancer exomes | 332 |
Query examples
COSMICMart allows data to be filtered on six different categories (Figure 1): cancer sample, gene, mutation, site of the tumour, histology and other (e.g. Ensembl Gene ID, Swissprot ID, Entrez Gene ID). The interface has a number of pre-selected filters and attributes; mutated samples are selected by default. Users can change these to suit their requirements. Results are displayed in tabulated form and are exportable in various formats for further analysis.
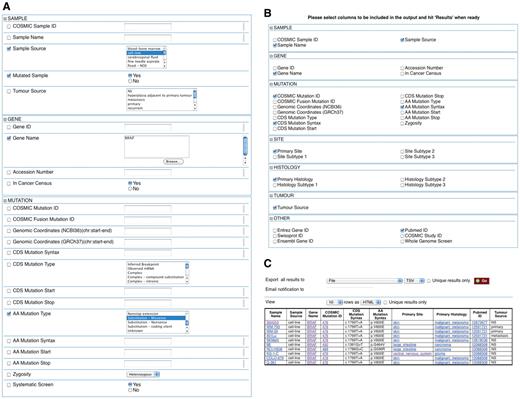
Example of how COSMICMart can be queried. This query searches for all cell lines with missense substitution mutations in the BRAF gene (A). Attributes can be selected (B) to display in the results table (C).
Query #1: ‘Find all missense substitution mutations for BRAF in cell lines, and display sample, mutation, site, and histology information’ (Figure 1, Table 2).
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Sample name |
| Sample source: cell-line | Sample source | |
| Gene name: BRAF | Gene name | |
| AA mutation type: substitution—missense | Cosmic mutation ID (COSM ID) | |
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Primary site | ||
| Primary histology | ||
| Tumour source | ||
| Pubmed ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Sample name |
| Sample source: cell-line | Sample source | |
| Gene name: BRAF | Gene name | |
| AA mutation type: substitution—missense | Cosmic mutation ID (COSM ID) | |
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Primary site | ||
| Primary histology | ||
| Tumour source | ||
| Pubmed ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Sample name |
| Sample source: cell-line | Sample source | |
| Gene name: BRAF | Gene name | |
| AA mutation type: substitution—missense | Cosmic mutation ID (COSM ID) | |
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Primary site | ||
| Primary histology | ||
| Tumour source | ||
| Pubmed ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Sample name |
| Sample source: cell-line | Sample source | |
| Gene name: BRAF | Gene name | |
| AA mutation type: substitution—missense | Cosmic mutation ID (COSM ID) | |
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Primary site | ||
| Primary histology | ||
| Tumour source | ||
| Pubmed ID |
Missense mutations are the most common variant type in COSMIC; over 90% of mutations in BRAF are missense mutations at the p.V600 position. The results are returned as a tabular summary with links back to the COSMIC website. The sample name field links back to the COSMIC sample overview web page, and mutation ID (COSM ID) to the COSMIC mutation summary page (Figure 2). From the COSMIC mutation summary web page, there are links to the Ensembl contig view so the mutation can be viewed in a genomic context. There are also links to the GMOD’s GBrowse where COSMIC coding and non-coding mutations, gene footprints, structural rearrangements and copy number variants can be viewed (11).
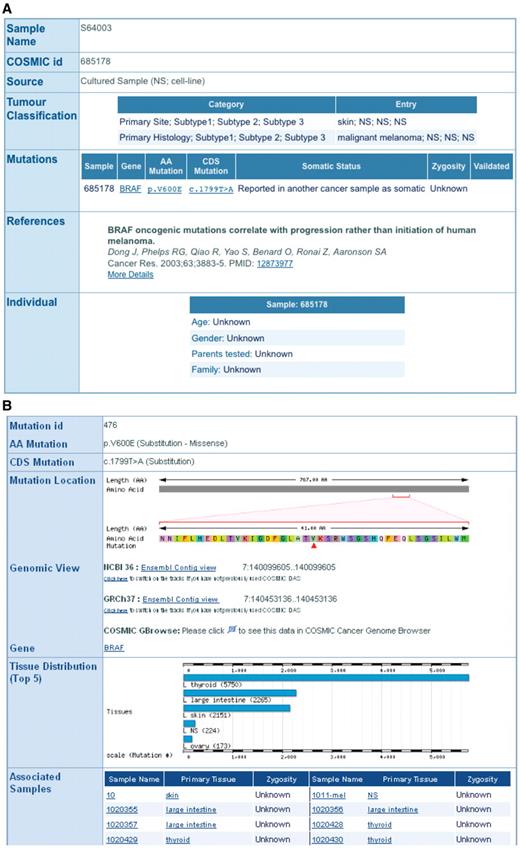
The COSMIC sample (A) and mutation (B) summary pages are linked directly from COSMICMart output table.
Query #2: ‘Find all gene fusion mutations involving the FUS gene with a primary site of bone, and display mutation and sample information’ (Table 3).
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Gene name: FUS | Sample name | |
| CDS mutation type: inferred breakpoint, observed mRNA | Sample source | |
| Primary site: bone | Cosmic fusion mutation ID | |
| CDS mutation syntax | ||
| Pubmed ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Gene name: FUS | Sample name | |
| CDS mutation type: inferred breakpoint, observed mRNA | Sample source | |
| Primary site: bone | Cosmic fusion mutation ID | |
| CDS mutation syntax | ||
| Pubmed ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Gene name: FUS | Sample name | |
| CDS mutation type: inferred breakpoint, observed mRNA | Sample source | |
| Primary site: bone | Cosmic fusion mutation ID | |
| CDS mutation syntax | ||
| Pubmed ID |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Gene name: FUS | Sample name | |
| CDS mutation type: inferred breakpoint, observed mRNA | Sample source | |
| Primary site: bone | Cosmic fusion mutation ID | |
| CDS mutation syntax | ||
| Pubmed ID |
Gene fusions have been associated with a number of specific tumour types including prostate and blood tumours. These biomarkers can be useful in diagnosis and as targets for drug therapies. COSMIC has annotations, for an increasing number of gene fusion mutations, which are viewable using COSMICMart. The COSMIC fusion mutation ID links to the gene fusion summary pages, which give a graphical view of different fusion structures observed. Many of the papers describing gene fusions have identified more than one gene fusion product for the same genes in a single sample. Observed mRNAs are the actual expressed products reported in the results. However, to aid display and website navigation, we have inferred the genomic breakpoint from the experimental data.
Query #3: ‘Find variation information in Ensembl for all genes from mutated samples with a primary site of breast, and display COSMIC gene, mutation and sample information along with Ensembl variation information’ (Table 4).
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Primary site: breast | Sample name | |
| Sample source | ||
| Cosmic mutation ID (COSM ID) | ||
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Ensembl: Homo sapiens genes | Features: Ensembl gene ID | |
| Features: Ensembl transcript ID | ||
| Variations: variation source | ||
| Variations: source description | ||
| Variations: reference ID | ||
| Variations: allele |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Primary site: breast | Sample name | |
| Sample source | ||
| Cosmic mutation ID (COSM ID) | ||
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Ensembl: Homo sapiens genes | Features: Ensembl gene ID | |
| Features: Ensembl transcript ID | ||
| Variations: variation source | ||
| Variations: source description | ||
| Variations: reference ID | ||
| Variations: allele |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Primary site: breast | Sample name | |
| Sample source | ||
| Cosmic mutation ID (COSM ID) | ||
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Ensembl: Homo sapiens genes | Features: Ensembl gene ID | |
| Features: Ensembl transcript ID | ||
| Variations: variation source | ||
| Variations: source description | ||
| Variations: reference ID | ||
| Variations: allele |
| Data sets . | Filters . | Attributes . |
|---|---|---|
| COSMIC51 | Mutated sample: yes | Cosmic sample ID |
| Primary site: breast | Sample name | |
| Sample source | ||
| Cosmic mutation ID (COSM ID) | ||
| CDS mutation syntax | ||
| AA mutation syntax | ||
| Ensembl: Homo sapiens genes | Features: Ensembl gene ID | |
| Features: Ensembl transcript ID | ||
| Variations: variation source | ||
| Variations: source description | ||
| Variations: reference ID | ||
| Variations: allele |
COSMICMart is federated with Ensembl (12), which allows Biomart queries to return and integrate data from both resources. For instance, the linking of the two resources can allow the retrieval of variation data from both resources (somatic mutations from COSMIC and germline polymorphisms from Ensembl) for a particular gene or set of genes or cancer type. There is an increasing awareness of how genomic variation can affect a tumour’s sensitivity or resistance to anti-cancer agents. While this genetic variation can be familial or somatic, an understanding of common genetic variation around known cancer genes can be of much value to investigations searching for loci modifying a tumour’s response to drug therapy (13–15). This query is achieved by first selecting the filters/attributes in the COSMIC BioMart and then clicking the ‘Dataset’ link at the bottom of the left hand margin of the BioMart interface. An additional data set can then be selected from the drop down list, in this instance Ensembl, which allows a federated query between COSMIC and Ensembl. The filters/attributes are then set in the usual way using the Ensembl BioMart to produce an integrated query.
Future directions
COSMIC will continue to curate newly discovered cancer genes and is committed to update existing cancer genes with a data release every 2 months. This will ensure that the scientific community has an up-to-date catalogue of somatic mutations implicated in human cancer. COSMICMart is also automatically updated with each new COSMIC release, which allows the data set to be easily mined and integrated with other resources. COSMIC has been successfully adapted to hold complete catalogues of somatic mutations for individual cancer samples. Currently COSMIC holds genome-wide data on 383 tumour samples and we expect this to increase in the near future.
It is intended to federate COSMICMart with further BioMart-driven data resources in addition to the current link with Ensembl. Linking our data to PRIDE (16), UniProt (17) and InterPro (18) will allow COSMIC somatic mutation data to be linked to protein and peptide annotation, while the addition of the Reactome (19) database will allow the incorporation of pathway data. We also intend to create direct links between COSMIC and the ICGC Data Portal (http://dcc.icgc.org/) so somatic mutation data can be integrated between the two data resources.
Funding
Funding for open access charge: Wellcome Trust (grant reference 077012/Z/05/Z).
Conflict of interest. None declared.



