-
PDF
- Split View
-
Views
-
Cite
Cite
Gengkon Lum, Xiang Jia Min, FunSecKB: the Fungal Secretome KnowledgeBase, Database, Volume 2011, 2011, bar001, https://doi.org/10.1093/database/bar001
Close - Share Icon Share
Abstract
The Fungal Secretome KnowledgeBase (FunSecKB) provides a resource of secreted fungal proteins, i.e. secretomes, identified from all available fungal protein data in the NCBI RefSeq database. The secreted proteins were identified using a well evaluated computational protocol which includes SignalP, WolfPsort and Phobius for signal peptide or subcellular location prediction, TMHMM for identifying membrane proteins, and PS-Scan for identifying endoplasmic reticulum (ER) target proteins. The entries were mapped to the UniProt database and any annotations of subcellular locations that were either manually curated or computationally predicted were included in FunSecKB. Using a web-based user interface, the database is searchable, browsable and downloadable by using NCBI’s RefSeq accession or gi number, UniProt accession number, keyword or by species. A BLAST utility was integrated to allow users to query the database by sequence similarity. A user submission tool was implemented to support community annotation of subcellular locations of fungal proteins. With the complete fungal data from RefSeq and associated web-based tools, FunSecKB will be a valuable resource for exploring the potential applications of fungal secreted proteins.
Database URL:http://proteomics.ysu.edu/secretomes/fungi.php
Introduction
Fungi play an important role in carbon cycling as they use secreted enzymes to break down lignocelluloses and other biopolymers then transporting the resulting products into the cells as their food. The secreted proteins in plant associated fungi play important roles in plant and fungi symbiosis or fungal pathogenicity (1). Fungal secreted proteins also play important roles in the development of fungal diseases in human (2,3). Secreted fungal enzymes have found a wide range of applications in the food, feed, pulp and paper, bioethanol and textile industries (4).
Signal-peptide dependent secreted proteins contain a signal peptide (SP) at the N-terminus that directs the ribosomes to the rough endoplasmic reticulum (ER) for completing polypeptide synthesis (5,6). The signal peptide, typically 15–30 amino acids long and consisting of 15–20 hydrophobic amino acid residues, is cleaved off during translocation across the membrane. While some proteins without an N-terminal signal peptide can be found in the ER and the Golgi, over 90% of human secreted proteins (7) and ∼90% of the Aspergillus niger extracellular proteins identified by mass spectrometry contain classical N-terminal signal peptides (8). There are also examples of non-classically secreted proteins in fungi, including the Saccharomyces cerevisiae mating pheromone a-factor (9) and two galectins from Coprinus cinereus (10), but it is generally believed that the vast majority of secreted fungal proteins are processed by the classical secretory pathway (8).
The term secretome is often used to refer to the complete set of secreted proteins in an organism (2,11,12). However, the term has also been used to include the set of proteins involved in the secretory pathway (13,14). In the work described here, the secretome only includes the secreted proteins in an organism. Along with an increased number of species having genomes being completely sequenced, we see an increased number of publications on fungal secretome identification and analysis using both computational and experimental approaches (15). For example, secretomes have been reported in following fungi including A. niger (8), Candida albicans (16), Phanerochaete chrysosporium (17), Sclerotinia sclerotiorum (18), Fusarium graminearum (19) and Ustilago maydis (20). Considering the biological importance of secreted proteins and their potential industrial applications, we developed a knowledgebase of fungal secretomes for identification, annotation and curation of both computationally predicted and experimentally identified fungal secreted proteins. This knowledgebase is designed to serve as a central portal for providing as well as collecting information on fungal secretomes.
Data collection and database implementation
The fungal protein sequences were retrieved from the NCBI Reference Sequence collection (RefSeq) database (release April, 2010) (http://www.ncbi.nlm.nih.gov/RefSeq/). The rational for choosing the RefSeq protein data set was that RefSeq provides a comprehensive, integrated, non-redundant, well-annotated set of proteins and also the corresponding nucleotide sequences were also linked for these protein sequences in their database (21). The data in the fungal secretome knowledgebase (FunSecKB) were obtained from the following three sources: (i) the features predicted using computational approaches; (ii) subcellular locations annotated in UniProtKB; and (iii) our manual curation with experimental evidence obtained from recent literature.
Computational methods for prediction of secreted proteins
The fungal protein sequences downloaded from the NCBI RefSeq database were processed using the following programs including SignalP (version 3.0, http://www.cbs.dtu.dk/services/SignalP/) (22), Phobius (http://phobius.binf.ku.dk/) (23,24), WolfPsort (http://wolfpsort.org/) (25,26) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) (27), for signal peptide and subcellular location prediction. We chose these four predictors because they were previously evaluated favorably and widely used by the fungal secretome research community (8,16,28). TMHMM (http://www.cbs.dtu.dk/services/TMHMM) was used to identify proteins having transmembrane domains (29) and PS-Scan (http://www.expasy.org/tools/scanprosite/) was used to scan ER targeting sequence (Prosite: PS00014) (30). With each of the programs, the default parameters for eukaryotes or fungi were used. For SignalP prediction, only entries that were predicted having a ‘mostly likely cleavage site’ by SignalP-NN algorithm and a ‘signal peptide’ by SignalP-HMM algorithm were considered to be true signal peptide ‘positives’, using the N-terminal 70 amino acids (22). For predicting membrane proteins using TMHMM, the entries having membrane domains not located within the N-terminus (the first 70 amino acids) were treated as real membrane proteins. Protein sequences predicted to have a signal peptide by SignalP were further processed using FragAnchor to identify the glycosylphosphatidyinositol (GPI) anchors (http://navet.ics.hawaii.edu/∼fraganchor/NNHMM/NNHMM.html) (31). Protein sequences predicted as having a GPI anchor may be attached to the outside of the plasma membrane or may be secreted to be targeted to the cell wall (32).
We recently performed the accuracy evaluation of the computational methods, using 241 experimentally identified secreted proteins and 5992 non-secreted proteins in fungi that were retrieved from UniProt/Swiss-Prot data set, and found that the highest prediction accuracy (92.1% in sensitivity and 98.9% in specificity) was achieved by combining SignalP, WolfPsort and Phobius for signal peptide prediction, TMHMM for eliminating membrane proteins, and PS-Scan for removing ER targeting proteins (28). Thus, the secretomes defined in this study include the manually curated secreted proteins along with the proteins predicted as having a signal peptide at their N-terminus by SignalP and Phobius and with a subcellular location predicted as extracellular by WolfPsort, but not having a transmembrane domain or an ER targeting signal. The information provided by TargetP and fragAnchor were also included in the annotation which may be useful for identifying mitochondrial targeted proteins or GPI anchored membrane or cell wall proteins. An overview of the database’s features are shown in Figure 1.
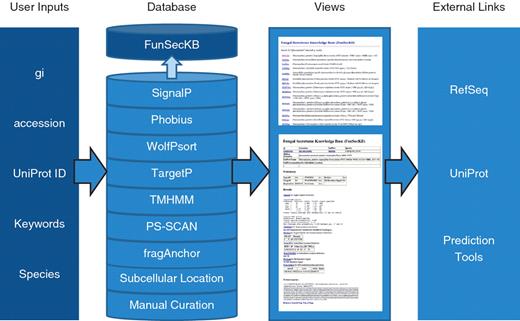
Overview of FunSecKB. To search the database users can enter NCBI RefSeq gi or accession number, UniProt accession number, keywords or species. The database consists of information generated using seven prediction tools and subcellular location annotated in UniProtKB and our own manual curation. Users can browse through the results using the web user-interface. Links to external databases and resources are also provided for further exploration. Whole secretome sequences can be downloaded and BLAST utility can be accessed from the database interface.
Linking RefSeq proteins to UniProtKB annotation
The fungal protein entries in FunSecKB are linked to the UniProtKB using the mapping information generated in UniProtKB (ftp://ftp.uniprot.org/pub/databases/uniprot/current release/knowledgebase/idmapping/) (33). We also integrated the subcellular location information of fungal proteins annotated in UniProtKB including curated (reviewed, from the UniProtKB/Swiss-Prot data set) and predicted (unreviewed, from the UniProtKB/TrEMBL data set). In addition, we also included manually curated protein entries in UniProtKB/Swiss-Prot data set which could not be mapped to entries in the RefSeq database.
Manual curation and community annotation
FunSecKB supports community curation of subcellular locations of fungal proteins based on published experimental evidence. A submission form was developed for users to provide subcellular location annotation and the literature source to support the annotation. After our curator’s validation, these data will be incorporated into the database. Currently we have manually curated more than two hundred secreted proteins from A. niger (8). Manual curation is an ongoing process, thus additional secreted proteins will be manually curated and integrated into the database with time.
The information from the above three sources are integrated in the annotation (Figure 1). The annotated entries are linked to the RefSeq database in NCBI and UniProtKB as well as related literature for entries manually curated by our curators or the community. The data will be updated when a new RefSeq data set is released from NCBI (http://www.ncbi.nlm.nih.gov/RefSeq/).
Data access
FunSecKB can be accessed through the database web interface at http://proteomics.ysu.edu/secretomes/fungi.php. There are three approaches to accessing the data including: (i) search individual proteins using NCBI’s RefSeq gi or accession number, UniProt accession number, keyword or by species; (ii) search or download the whole secretome or a subset of manually curated secreted proteins of a species and (iii) search all fungal proteins or fungal secreted proteins using BLAST.
The annotation page contains the summary and the details of subcellular locations predicted by the tools mentioned above and annotation retrieved from UniProtKB. Each entry is linked to both RefSeq and UniProtKB. The secretome, including predicted and curated secreted proteins from a particular species, can be searched and downloaded by selecting a species from the species list for complete genomes or inputting a species name for others not having a complete genome. The protein sequences of the secretome from a species can be downloaded into a fasta file. Manually curated secreted proteins consist of proteins retrieved from UniProtKB/Swiss-Prot with subcellular locations labeled as ‘reviewed’ and proteins curated by our curators and the users. The proteins curated by us and by the community are supported by experimental evidence for their subcellular location annotation and the related literature can be found on the same page. The annotation page also contains the primary protein sequence (Figure 1). The database interface provides a link to the BLAST input interface to search through the proteins retrieved from RefSeq: either all fungal proteins or just the fungal secretomes.
Preliminary data analysis
Currently FunSecKB contains a total of 478 073 fungal protein sequences including 23 878 predicted and/or curated secreted proteins from a total of 118 fungal species. This includes 52 fungal species, with one species having two different varieties, having a complete predicted proteome set. We performed a preliminary analysis on the 53 complete secretomes of 52 fungal species including 43 Ascomycetes, 7 Basidiomycetes (with Cryptococcus neoformans having two varieties) and 2 Microsporidia (Table 1). Overall, fungal species having an expanded genome size encode more proteins in their predicted proteomes (r = 0.75) (Figure 2a). Ajellomyces dermatitidis and Postia placenta are two outliers. For the P. placenta genome of 69 Mb the RefSeq only has 9083 predicted proteins, however, Martinez et al. (2009) reported 17 173 proteins predicted from the P. placenta genome (34). Thus the discrepancy may be caused by lagged database update. The reason for the A. dermatitidis data is not known.
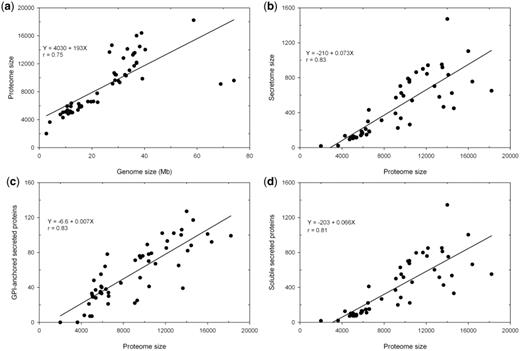
Relationship between genome size, proteome size and secretome size in fungi. (a) genome size and proteome size; (b) proteome size and secretome size; (c) proteome size and GPI-anchored secreted proteins and (d) proteome size and soluble secreted proteins.
| Species . | Phylum . | Genome (Mb) . | Predicted Proteome . | Predicted Secretome . | Curated Secretome . | GPI-anchored Secretome . | Soluble Secretome . | Secretome (%) . | GPI-anchored Portion (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Ajellomyces capsulatus | Ascomycota | 31 | 9313 | 224 | 0 | 25 | 199 | 2.4 | 11.2 |
| Ajellomyces dermatitidis | Ascomycota | 74 | 9587 | 335 | 0 | 51 | 284 | 3.5 | 15.2 |
| Ashbya gossypii | Ascomycota | 8 | 4725 | 93 | 2 | 21 | 72 | 2.0 | 22.6 |
| Aspergillus clavatus | Ascomycota | 28 | 9121 | 571 | 17 | 71 | 500 | 6.3 | 12.4 |
| Aspergillus flavus | Ascomycota | 36 | 13 487 | 951 | 25 | 100 | 851 | 7.1 | 10.5 |
| Aspergillus fumigatus | Ascomycota | 29 | 9630 | 624 | 58 | 74 | 550 | 6.5 | 11.9 |
| Aspergillus nidulans | Ascomycota | 30 | 9541 | 704 | 29 | 76 | 628 | 7.4 | 10.8 |
| Aspergillus niger | Ascomycota | 34 | 14 102 | 832 | 253 | 82 | 750 | 5.9 | 9.9 |
| Aspergillus oryzae | Ascomycota | 37 | 12 074 | 843 | 28 | 85 | 758 | 7.0 | 10.1 |
| Aspergillus terreus | Ascomycota | 29 | 10 401 | 774 | 23 | 70 | 704 | 7.4 | 9.0 |
| Botryotinia fuckeliana | Ascomycota | 39 | 16 389 | 755 | 4 | 92 | 663 | 4.6 | 12.2 |
| Candida albicans | Ascomycota | 28 | 14 633 | 449 | 41 | 117 | 332 | 3.1 | 26.1 |
| Candida dubliniensis | Ascomycota | 16 | 5860 | 184 | 0 | 55 | 129 | 3.1 | 29.9 |
| Candida glabrata | Ascomycota | 12 | 5192 | 121 | 7 | 48 | 73 | 2.3 | 39.7 |
| Candida tropicalis | Ascomycota | 15 | 6254 | 212 | 1 | 64 | 148 | 3.4 | 30.2 |
| Chaetomium globosum | Ascomycota | 34 | 11 048 | 862 | 1 | 67 | 795 | 7.8 | 7.8 |
| Clavispora lusitaniae | Ascomycota | 16 | 5936 | 169 | 0 | 40 | 129 | 2.8 | 23.7 |
| Coccidioides immitis | Ascomycota | 29 | 10 440 | 263 | 2 | 41 | 222 | 2.5 | 15.6 |
| Debaryomyces hansenii | Ascomycota | 12 | 6335 | 148 | 1 | 38 | 110 | 2.3 | 25.7 |
| Gibberella zeae | Ascomycota | 36 | 11 690 | 900 | 1 | 102 | 798 | 7.7 | 11.3 |
| Kluyveromyces lactis | Ascomycota | 11 | 5357 | 113 | 5 | 37 | 76 | 2.1 | 32.7 |
| Lachancea thermotolerans | Ascomycota | 10 | 5091 | 128 | 0 | 29 | 99 | 2.5 | 22.7 |
| Lodderomyces elongisporus | Ascomycota | 16 | 5799 | 139 | 0 | 34 | 105 | 2.4 | 24.5 |
| Magnaporthe grisea | Ascomycota | 40 | 14 010 | 1471 | 3 | 127 | 1344 | 10.5 | 8.6 |
| Neosartorya fischeri | Ascomycota | 33 | 10 406 | 751 | 21 | 78 | 673 | 7.2 | 10.4 |
| Neurospora crassa | Ascomycota | 39 | 9844 | 592 | 10 | 76 | 516 | 6.0 | 12.8 |
| Penicillium chrysogenum | Ascomycota | 32 | 12 791 | 703 | 5 | 102 | 601 | 5.5 | 14.5 |
| Penicillium marneffei | Ascomycota | 29 | 10 663 | 538 | 0 | 79 | 459 | 5.0 | 14.7 |
| Phaeosphaeria nodorum | Ascomycota | 37 | 16 002 | 1103 | 1 | 101 | 1002 | 6.9 | 9.2 |
| Pichia guilliermondii | Ascomycota | 11 | 5920 | 159 | 0 | 33 | 126 | 2.7 | 20.8 |
| Pichia pastoris | Ascomycota | 9 | 5040 | 105 | 0 | 31 | 74 | 2.1 | 29.5 |
| Pichia stipitis | Ascomycota | 15 | 5816 | 144 | 0 | 35 | 109 | 2.5 | 24.3 |
| Podospora anserina | Ascomycota | 33 | 10 272 | 789 | 1 | 89 | 700 | 7.7 | 11.3 |
| Pyrenophora tritici-repentis | Ascomycota | 37 | 12 169 | 942 | 0 | 93 | 849 | 7.7 | 9.9 |
| Saccharomyces cerevisiae | Ascomycota | 12 | 5885 | 156 | 101 | 41 | 115 | 2.7 | 26.3 |
| Schizosaccharomyces japonicus | Ascomycota | 11 | 4824 | 109 | 0 | 7 | 102 | 2.3 | 6.4 |
| Schizosaccharomyces pombe | Ascomycota | 13 | 5001 | 112 | 43 | 7 | 105 | 2.2 | 6.3 |
| Sclerotinia sclerotiorum | Ascomycota | 38 | 14 446 | 623 | 1 | 88 | 535 | 4.3 | 14.1 |
| Talaromyces stipitatus | Ascomycota | 36 | 13 252 | 580 | 0 | 65 | 515 | 4.4 | 11.2 |
| Uncinocarpus reesii | Ascomycota | 22 | 7760 | 312 | 0 | 45 | 267 | 4.0 | 14.4 |
| Vanderwaltozyma polyspora | Ascomycota | 15 | 5376 | 116 | 0 | 28 | 88 | 2.2 | 24.1 |
| Yarrowia lipolytica | Ascomycota | 22 | 6472 | 299 | 5 | 78 | 221 | 4.6 | 26.1 |
| Zygosaccharomyces rouxii | Ascomycota | 12 | 4994 | 120 | 0 | 33 | 87 | 2.4 | 27.5 |
| Coprinopsis cinerea | Basidiomycota | 36 | 13 546 | 917 | 8 | 106 | 811 | 6.8 | 11.6 |
| Cryptococcus neoformans (neoformans B-3501A) | Basidiomycota | 19 | 6578 | 186 | 0 | 34 | 152 | 2.8 | 18.3 |
| Cryptococcus neoformans (neoformans JEC21) | Basidiomycota | 21 | 6594 | 181 | 0 | 30 | 151 | 2.7 | 16.6 |
| Laccaria bicolor | Basidiomycota | 59 | 18 215 | 650 | 0 | 99 | 551 | 3.6 | 15.2 |
| Malassezia globosa | Basidiomycota | 9 | 4286 | 134 | 0 | 8 | 126 | 3.1 | 6.0 |
| Moniliophthora perniciosa | Basidiomycota | 27 | 13 649 | 465 | 0 | 39 | 426 | 3.4 | 8.4 |
| Postia placenta | Basidiomycota | 69 | 9083 | 391 | 0 | 22 | 369 | 4.3 | 5.6 |
| Ustilago maydis | Basidiomycota | 20 | 6548 | 431 | 2 | 21 | 410 | 6.6 | 4.9 |
| Encephalitozoon cuniculi | Microsporidia | 3 | 1996 | 17 | 2 | 0 | 17 | 0.9 | 0.0 |
| Enterocytozoon bieneusi | Microsporidia | 4 | 3632 | 21 | 0 | 0 | 21 | 0.6 | 0.0 |
| Other species | 998 | 367 | 366 | ||||||
| Total | 478 073 | 23 878 | 1067 | 3014 |
| Species . | Phylum . | Genome (Mb) . | Predicted Proteome . | Predicted Secretome . | Curated Secretome . | GPI-anchored Secretome . | Soluble Secretome . | Secretome (%) . | GPI-anchored Portion (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Ajellomyces capsulatus | Ascomycota | 31 | 9313 | 224 | 0 | 25 | 199 | 2.4 | 11.2 |
| Ajellomyces dermatitidis | Ascomycota | 74 | 9587 | 335 | 0 | 51 | 284 | 3.5 | 15.2 |
| Ashbya gossypii | Ascomycota | 8 | 4725 | 93 | 2 | 21 | 72 | 2.0 | 22.6 |
| Aspergillus clavatus | Ascomycota | 28 | 9121 | 571 | 17 | 71 | 500 | 6.3 | 12.4 |
| Aspergillus flavus | Ascomycota | 36 | 13 487 | 951 | 25 | 100 | 851 | 7.1 | 10.5 |
| Aspergillus fumigatus | Ascomycota | 29 | 9630 | 624 | 58 | 74 | 550 | 6.5 | 11.9 |
| Aspergillus nidulans | Ascomycota | 30 | 9541 | 704 | 29 | 76 | 628 | 7.4 | 10.8 |
| Aspergillus niger | Ascomycota | 34 | 14 102 | 832 | 253 | 82 | 750 | 5.9 | 9.9 |
| Aspergillus oryzae | Ascomycota | 37 | 12 074 | 843 | 28 | 85 | 758 | 7.0 | 10.1 |
| Aspergillus terreus | Ascomycota | 29 | 10 401 | 774 | 23 | 70 | 704 | 7.4 | 9.0 |
| Botryotinia fuckeliana | Ascomycota | 39 | 16 389 | 755 | 4 | 92 | 663 | 4.6 | 12.2 |
| Candida albicans | Ascomycota | 28 | 14 633 | 449 | 41 | 117 | 332 | 3.1 | 26.1 |
| Candida dubliniensis | Ascomycota | 16 | 5860 | 184 | 0 | 55 | 129 | 3.1 | 29.9 |
| Candida glabrata | Ascomycota | 12 | 5192 | 121 | 7 | 48 | 73 | 2.3 | 39.7 |
| Candida tropicalis | Ascomycota | 15 | 6254 | 212 | 1 | 64 | 148 | 3.4 | 30.2 |
| Chaetomium globosum | Ascomycota | 34 | 11 048 | 862 | 1 | 67 | 795 | 7.8 | 7.8 |
| Clavispora lusitaniae | Ascomycota | 16 | 5936 | 169 | 0 | 40 | 129 | 2.8 | 23.7 |
| Coccidioides immitis | Ascomycota | 29 | 10 440 | 263 | 2 | 41 | 222 | 2.5 | 15.6 |
| Debaryomyces hansenii | Ascomycota | 12 | 6335 | 148 | 1 | 38 | 110 | 2.3 | 25.7 |
| Gibberella zeae | Ascomycota | 36 | 11 690 | 900 | 1 | 102 | 798 | 7.7 | 11.3 |
| Kluyveromyces lactis | Ascomycota | 11 | 5357 | 113 | 5 | 37 | 76 | 2.1 | 32.7 |
| Lachancea thermotolerans | Ascomycota | 10 | 5091 | 128 | 0 | 29 | 99 | 2.5 | 22.7 |
| Lodderomyces elongisporus | Ascomycota | 16 | 5799 | 139 | 0 | 34 | 105 | 2.4 | 24.5 |
| Magnaporthe grisea | Ascomycota | 40 | 14 010 | 1471 | 3 | 127 | 1344 | 10.5 | 8.6 |
| Neosartorya fischeri | Ascomycota | 33 | 10 406 | 751 | 21 | 78 | 673 | 7.2 | 10.4 |
| Neurospora crassa | Ascomycota | 39 | 9844 | 592 | 10 | 76 | 516 | 6.0 | 12.8 |
| Penicillium chrysogenum | Ascomycota | 32 | 12 791 | 703 | 5 | 102 | 601 | 5.5 | 14.5 |
| Penicillium marneffei | Ascomycota | 29 | 10 663 | 538 | 0 | 79 | 459 | 5.0 | 14.7 |
| Phaeosphaeria nodorum | Ascomycota | 37 | 16 002 | 1103 | 1 | 101 | 1002 | 6.9 | 9.2 |
| Pichia guilliermondii | Ascomycota | 11 | 5920 | 159 | 0 | 33 | 126 | 2.7 | 20.8 |
| Pichia pastoris | Ascomycota | 9 | 5040 | 105 | 0 | 31 | 74 | 2.1 | 29.5 |
| Pichia stipitis | Ascomycota | 15 | 5816 | 144 | 0 | 35 | 109 | 2.5 | 24.3 |
| Podospora anserina | Ascomycota | 33 | 10 272 | 789 | 1 | 89 | 700 | 7.7 | 11.3 |
| Pyrenophora tritici-repentis | Ascomycota | 37 | 12 169 | 942 | 0 | 93 | 849 | 7.7 | 9.9 |
| Saccharomyces cerevisiae | Ascomycota | 12 | 5885 | 156 | 101 | 41 | 115 | 2.7 | 26.3 |
| Schizosaccharomyces japonicus | Ascomycota | 11 | 4824 | 109 | 0 | 7 | 102 | 2.3 | 6.4 |
| Schizosaccharomyces pombe | Ascomycota | 13 | 5001 | 112 | 43 | 7 | 105 | 2.2 | 6.3 |
| Sclerotinia sclerotiorum | Ascomycota | 38 | 14 446 | 623 | 1 | 88 | 535 | 4.3 | 14.1 |
| Talaromyces stipitatus | Ascomycota | 36 | 13 252 | 580 | 0 | 65 | 515 | 4.4 | 11.2 |
| Uncinocarpus reesii | Ascomycota | 22 | 7760 | 312 | 0 | 45 | 267 | 4.0 | 14.4 |
| Vanderwaltozyma polyspora | Ascomycota | 15 | 5376 | 116 | 0 | 28 | 88 | 2.2 | 24.1 |
| Yarrowia lipolytica | Ascomycota | 22 | 6472 | 299 | 5 | 78 | 221 | 4.6 | 26.1 |
| Zygosaccharomyces rouxii | Ascomycota | 12 | 4994 | 120 | 0 | 33 | 87 | 2.4 | 27.5 |
| Coprinopsis cinerea | Basidiomycota | 36 | 13 546 | 917 | 8 | 106 | 811 | 6.8 | 11.6 |
| Cryptococcus neoformans (neoformans B-3501A) | Basidiomycota | 19 | 6578 | 186 | 0 | 34 | 152 | 2.8 | 18.3 |
| Cryptococcus neoformans (neoformans JEC21) | Basidiomycota | 21 | 6594 | 181 | 0 | 30 | 151 | 2.7 | 16.6 |
| Laccaria bicolor | Basidiomycota | 59 | 18 215 | 650 | 0 | 99 | 551 | 3.6 | 15.2 |
| Malassezia globosa | Basidiomycota | 9 | 4286 | 134 | 0 | 8 | 126 | 3.1 | 6.0 |
| Moniliophthora perniciosa | Basidiomycota | 27 | 13 649 | 465 | 0 | 39 | 426 | 3.4 | 8.4 |
| Postia placenta | Basidiomycota | 69 | 9083 | 391 | 0 | 22 | 369 | 4.3 | 5.6 |
| Ustilago maydis | Basidiomycota | 20 | 6548 | 431 | 2 | 21 | 410 | 6.6 | 4.9 |
| Encephalitozoon cuniculi | Microsporidia | 3 | 1996 | 17 | 2 | 0 | 17 | 0.9 | 0.0 |
| Enterocytozoon bieneusi | Microsporidia | 4 | 3632 | 21 | 0 | 0 | 21 | 0.6 | 0.0 |
| Other species | 998 | 367 | 366 | ||||||
| Total | 478 073 | 23 878 | 1067 | 3014 |
| Species . | Phylum . | Genome (Mb) . | Predicted Proteome . | Predicted Secretome . | Curated Secretome . | GPI-anchored Secretome . | Soluble Secretome . | Secretome (%) . | GPI-anchored Portion (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Ajellomyces capsulatus | Ascomycota | 31 | 9313 | 224 | 0 | 25 | 199 | 2.4 | 11.2 |
| Ajellomyces dermatitidis | Ascomycota | 74 | 9587 | 335 | 0 | 51 | 284 | 3.5 | 15.2 |
| Ashbya gossypii | Ascomycota | 8 | 4725 | 93 | 2 | 21 | 72 | 2.0 | 22.6 |
| Aspergillus clavatus | Ascomycota | 28 | 9121 | 571 | 17 | 71 | 500 | 6.3 | 12.4 |
| Aspergillus flavus | Ascomycota | 36 | 13 487 | 951 | 25 | 100 | 851 | 7.1 | 10.5 |
| Aspergillus fumigatus | Ascomycota | 29 | 9630 | 624 | 58 | 74 | 550 | 6.5 | 11.9 |
| Aspergillus nidulans | Ascomycota | 30 | 9541 | 704 | 29 | 76 | 628 | 7.4 | 10.8 |
| Aspergillus niger | Ascomycota | 34 | 14 102 | 832 | 253 | 82 | 750 | 5.9 | 9.9 |
| Aspergillus oryzae | Ascomycota | 37 | 12 074 | 843 | 28 | 85 | 758 | 7.0 | 10.1 |
| Aspergillus terreus | Ascomycota | 29 | 10 401 | 774 | 23 | 70 | 704 | 7.4 | 9.0 |
| Botryotinia fuckeliana | Ascomycota | 39 | 16 389 | 755 | 4 | 92 | 663 | 4.6 | 12.2 |
| Candida albicans | Ascomycota | 28 | 14 633 | 449 | 41 | 117 | 332 | 3.1 | 26.1 |
| Candida dubliniensis | Ascomycota | 16 | 5860 | 184 | 0 | 55 | 129 | 3.1 | 29.9 |
| Candida glabrata | Ascomycota | 12 | 5192 | 121 | 7 | 48 | 73 | 2.3 | 39.7 |
| Candida tropicalis | Ascomycota | 15 | 6254 | 212 | 1 | 64 | 148 | 3.4 | 30.2 |
| Chaetomium globosum | Ascomycota | 34 | 11 048 | 862 | 1 | 67 | 795 | 7.8 | 7.8 |
| Clavispora lusitaniae | Ascomycota | 16 | 5936 | 169 | 0 | 40 | 129 | 2.8 | 23.7 |
| Coccidioides immitis | Ascomycota | 29 | 10 440 | 263 | 2 | 41 | 222 | 2.5 | 15.6 |
| Debaryomyces hansenii | Ascomycota | 12 | 6335 | 148 | 1 | 38 | 110 | 2.3 | 25.7 |
| Gibberella zeae | Ascomycota | 36 | 11 690 | 900 | 1 | 102 | 798 | 7.7 | 11.3 |
| Kluyveromyces lactis | Ascomycota | 11 | 5357 | 113 | 5 | 37 | 76 | 2.1 | 32.7 |
| Lachancea thermotolerans | Ascomycota | 10 | 5091 | 128 | 0 | 29 | 99 | 2.5 | 22.7 |
| Lodderomyces elongisporus | Ascomycota | 16 | 5799 | 139 | 0 | 34 | 105 | 2.4 | 24.5 |
| Magnaporthe grisea | Ascomycota | 40 | 14 010 | 1471 | 3 | 127 | 1344 | 10.5 | 8.6 |
| Neosartorya fischeri | Ascomycota | 33 | 10 406 | 751 | 21 | 78 | 673 | 7.2 | 10.4 |
| Neurospora crassa | Ascomycota | 39 | 9844 | 592 | 10 | 76 | 516 | 6.0 | 12.8 |
| Penicillium chrysogenum | Ascomycota | 32 | 12 791 | 703 | 5 | 102 | 601 | 5.5 | 14.5 |
| Penicillium marneffei | Ascomycota | 29 | 10 663 | 538 | 0 | 79 | 459 | 5.0 | 14.7 |
| Phaeosphaeria nodorum | Ascomycota | 37 | 16 002 | 1103 | 1 | 101 | 1002 | 6.9 | 9.2 |
| Pichia guilliermondii | Ascomycota | 11 | 5920 | 159 | 0 | 33 | 126 | 2.7 | 20.8 |
| Pichia pastoris | Ascomycota | 9 | 5040 | 105 | 0 | 31 | 74 | 2.1 | 29.5 |
| Pichia stipitis | Ascomycota | 15 | 5816 | 144 | 0 | 35 | 109 | 2.5 | 24.3 |
| Podospora anserina | Ascomycota | 33 | 10 272 | 789 | 1 | 89 | 700 | 7.7 | 11.3 |
| Pyrenophora tritici-repentis | Ascomycota | 37 | 12 169 | 942 | 0 | 93 | 849 | 7.7 | 9.9 |
| Saccharomyces cerevisiae | Ascomycota | 12 | 5885 | 156 | 101 | 41 | 115 | 2.7 | 26.3 |
| Schizosaccharomyces japonicus | Ascomycota | 11 | 4824 | 109 | 0 | 7 | 102 | 2.3 | 6.4 |
| Schizosaccharomyces pombe | Ascomycota | 13 | 5001 | 112 | 43 | 7 | 105 | 2.2 | 6.3 |
| Sclerotinia sclerotiorum | Ascomycota | 38 | 14 446 | 623 | 1 | 88 | 535 | 4.3 | 14.1 |
| Talaromyces stipitatus | Ascomycota | 36 | 13 252 | 580 | 0 | 65 | 515 | 4.4 | 11.2 |
| Uncinocarpus reesii | Ascomycota | 22 | 7760 | 312 | 0 | 45 | 267 | 4.0 | 14.4 |
| Vanderwaltozyma polyspora | Ascomycota | 15 | 5376 | 116 | 0 | 28 | 88 | 2.2 | 24.1 |
| Yarrowia lipolytica | Ascomycota | 22 | 6472 | 299 | 5 | 78 | 221 | 4.6 | 26.1 |
| Zygosaccharomyces rouxii | Ascomycota | 12 | 4994 | 120 | 0 | 33 | 87 | 2.4 | 27.5 |
| Coprinopsis cinerea | Basidiomycota | 36 | 13 546 | 917 | 8 | 106 | 811 | 6.8 | 11.6 |
| Cryptococcus neoformans (neoformans B-3501A) | Basidiomycota | 19 | 6578 | 186 | 0 | 34 | 152 | 2.8 | 18.3 |
| Cryptococcus neoformans (neoformans JEC21) | Basidiomycota | 21 | 6594 | 181 | 0 | 30 | 151 | 2.7 | 16.6 |
| Laccaria bicolor | Basidiomycota | 59 | 18 215 | 650 | 0 | 99 | 551 | 3.6 | 15.2 |
| Malassezia globosa | Basidiomycota | 9 | 4286 | 134 | 0 | 8 | 126 | 3.1 | 6.0 |
| Moniliophthora perniciosa | Basidiomycota | 27 | 13 649 | 465 | 0 | 39 | 426 | 3.4 | 8.4 |
| Postia placenta | Basidiomycota | 69 | 9083 | 391 | 0 | 22 | 369 | 4.3 | 5.6 |
| Ustilago maydis | Basidiomycota | 20 | 6548 | 431 | 2 | 21 | 410 | 6.6 | 4.9 |
| Encephalitozoon cuniculi | Microsporidia | 3 | 1996 | 17 | 2 | 0 | 17 | 0.9 | 0.0 |
| Enterocytozoon bieneusi | Microsporidia | 4 | 3632 | 21 | 0 | 0 | 21 | 0.6 | 0.0 |
| Other species | 998 | 367 | 366 | ||||||
| Total | 478 073 | 23 878 | 1067 | 3014 |
| Species . | Phylum . | Genome (Mb) . | Predicted Proteome . | Predicted Secretome . | Curated Secretome . | GPI-anchored Secretome . | Soluble Secretome . | Secretome (%) . | GPI-anchored Portion (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Ajellomyces capsulatus | Ascomycota | 31 | 9313 | 224 | 0 | 25 | 199 | 2.4 | 11.2 |
| Ajellomyces dermatitidis | Ascomycota | 74 | 9587 | 335 | 0 | 51 | 284 | 3.5 | 15.2 |
| Ashbya gossypii | Ascomycota | 8 | 4725 | 93 | 2 | 21 | 72 | 2.0 | 22.6 |
| Aspergillus clavatus | Ascomycota | 28 | 9121 | 571 | 17 | 71 | 500 | 6.3 | 12.4 |
| Aspergillus flavus | Ascomycota | 36 | 13 487 | 951 | 25 | 100 | 851 | 7.1 | 10.5 |
| Aspergillus fumigatus | Ascomycota | 29 | 9630 | 624 | 58 | 74 | 550 | 6.5 | 11.9 |
| Aspergillus nidulans | Ascomycota | 30 | 9541 | 704 | 29 | 76 | 628 | 7.4 | 10.8 |
| Aspergillus niger | Ascomycota | 34 | 14 102 | 832 | 253 | 82 | 750 | 5.9 | 9.9 |
| Aspergillus oryzae | Ascomycota | 37 | 12 074 | 843 | 28 | 85 | 758 | 7.0 | 10.1 |
| Aspergillus terreus | Ascomycota | 29 | 10 401 | 774 | 23 | 70 | 704 | 7.4 | 9.0 |
| Botryotinia fuckeliana | Ascomycota | 39 | 16 389 | 755 | 4 | 92 | 663 | 4.6 | 12.2 |
| Candida albicans | Ascomycota | 28 | 14 633 | 449 | 41 | 117 | 332 | 3.1 | 26.1 |
| Candida dubliniensis | Ascomycota | 16 | 5860 | 184 | 0 | 55 | 129 | 3.1 | 29.9 |
| Candida glabrata | Ascomycota | 12 | 5192 | 121 | 7 | 48 | 73 | 2.3 | 39.7 |
| Candida tropicalis | Ascomycota | 15 | 6254 | 212 | 1 | 64 | 148 | 3.4 | 30.2 |
| Chaetomium globosum | Ascomycota | 34 | 11 048 | 862 | 1 | 67 | 795 | 7.8 | 7.8 |
| Clavispora lusitaniae | Ascomycota | 16 | 5936 | 169 | 0 | 40 | 129 | 2.8 | 23.7 |
| Coccidioides immitis | Ascomycota | 29 | 10 440 | 263 | 2 | 41 | 222 | 2.5 | 15.6 |
| Debaryomyces hansenii | Ascomycota | 12 | 6335 | 148 | 1 | 38 | 110 | 2.3 | 25.7 |
| Gibberella zeae | Ascomycota | 36 | 11 690 | 900 | 1 | 102 | 798 | 7.7 | 11.3 |
| Kluyveromyces lactis | Ascomycota | 11 | 5357 | 113 | 5 | 37 | 76 | 2.1 | 32.7 |
| Lachancea thermotolerans | Ascomycota | 10 | 5091 | 128 | 0 | 29 | 99 | 2.5 | 22.7 |
| Lodderomyces elongisporus | Ascomycota | 16 | 5799 | 139 | 0 | 34 | 105 | 2.4 | 24.5 |
| Magnaporthe grisea | Ascomycota | 40 | 14 010 | 1471 | 3 | 127 | 1344 | 10.5 | 8.6 |
| Neosartorya fischeri | Ascomycota | 33 | 10 406 | 751 | 21 | 78 | 673 | 7.2 | 10.4 |
| Neurospora crassa | Ascomycota | 39 | 9844 | 592 | 10 | 76 | 516 | 6.0 | 12.8 |
| Penicillium chrysogenum | Ascomycota | 32 | 12 791 | 703 | 5 | 102 | 601 | 5.5 | 14.5 |
| Penicillium marneffei | Ascomycota | 29 | 10 663 | 538 | 0 | 79 | 459 | 5.0 | 14.7 |
| Phaeosphaeria nodorum | Ascomycota | 37 | 16 002 | 1103 | 1 | 101 | 1002 | 6.9 | 9.2 |
| Pichia guilliermondii | Ascomycota | 11 | 5920 | 159 | 0 | 33 | 126 | 2.7 | 20.8 |
| Pichia pastoris | Ascomycota | 9 | 5040 | 105 | 0 | 31 | 74 | 2.1 | 29.5 |
| Pichia stipitis | Ascomycota | 15 | 5816 | 144 | 0 | 35 | 109 | 2.5 | 24.3 |
| Podospora anserina | Ascomycota | 33 | 10 272 | 789 | 1 | 89 | 700 | 7.7 | 11.3 |
| Pyrenophora tritici-repentis | Ascomycota | 37 | 12 169 | 942 | 0 | 93 | 849 | 7.7 | 9.9 |
| Saccharomyces cerevisiae | Ascomycota | 12 | 5885 | 156 | 101 | 41 | 115 | 2.7 | 26.3 |
| Schizosaccharomyces japonicus | Ascomycota | 11 | 4824 | 109 | 0 | 7 | 102 | 2.3 | 6.4 |
| Schizosaccharomyces pombe | Ascomycota | 13 | 5001 | 112 | 43 | 7 | 105 | 2.2 | 6.3 |
| Sclerotinia sclerotiorum | Ascomycota | 38 | 14 446 | 623 | 1 | 88 | 535 | 4.3 | 14.1 |
| Talaromyces stipitatus | Ascomycota | 36 | 13 252 | 580 | 0 | 65 | 515 | 4.4 | 11.2 |
| Uncinocarpus reesii | Ascomycota | 22 | 7760 | 312 | 0 | 45 | 267 | 4.0 | 14.4 |
| Vanderwaltozyma polyspora | Ascomycota | 15 | 5376 | 116 | 0 | 28 | 88 | 2.2 | 24.1 |
| Yarrowia lipolytica | Ascomycota | 22 | 6472 | 299 | 5 | 78 | 221 | 4.6 | 26.1 |
| Zygosaccharomyces rouxii | Ascomycota | 12 | 4994 | 120 | 0 | 33 | 87 | 2.4 | 27.5 |
| Coprinopsis cinerea | Basidiomycota | 36 | 13 546 | 917 | 8 | 106 | 811 | 6.8 | 11.6 |
| Cryptococcus neoformans (neoformans B-3501A) | Basidiomycota | 19 | 6578 | 186 | 0 | 34 | 152 | 2.8 | 18.3 |
| Cryptococcus neoformans (neoformans JEC21) | Basidiomycota | 21 | 6594 | 181 | 0 | 30 | 151 | 2.7 | 16.6 |
| Laccaria bicolor | Basidiomycota | 59 | 18 215 | 650 | 0 | 99 | 551 | 3.6 | 15.2 |
| Malassezia globosa | Basidiomycota | 9 | 4286 | 134 | 0 | 8 | 126 | 3.1 | 6.0 |
| Moniliophthora perniciosa | Basidiomycota | 27 | 13 649 | 465 | 0 | 39 | 426 | 3.4 | 8.4 |
| Postia placenta | Basidiomycota | 69 | 9083 | 391 | 0 | 22 | 369 | 4.3 | 5.6 |
| Ustilago maydis | Basidiomycota | 20 | 6548 | 431 | 2 | 21 | 410 | 6.6 | 4.9 |
| Encephalitozoon cuniculi | Microsporidia | 3 | 1996 | 17 | 2 | 0 | 17 | 0.9 | 0.0 |
| Enterocytozoon bieneusi | Microsporidia | 4 | 3632 | 21 | 0 | 0 | 21 | 0.6 | 0.0 |
| Other species | 998 | 367 | 366 | ||||||
| Total | 478 073 | 23 878 | 1067 | 3014 |
The proportion of the secretomes in the proteomes in different species varies significantly from <1% in Encephalitozoon cuniculi and Enterocytozoon bieneusi, two Microsporidia species (unicellular parasites), to >10% in Magnaporthe grisea, a rice pathogenic fungus (Table 1). Overall, predicted secretome sizes increase with expanded proteome sizes in fungal species (r = 0.83) (Figure 2b). We further identified GPI-anchored proteins in the predicted secretome, which represent insoluble portions of secreted proteins that are components of cell walls or attached to the outside of cell membrane. We see that both insoluble and soluble portions are increased with increased proteome size in different fungal species (Figure 2c and 2d).
The functional categorization of predicted secretomes was analyzed using the rpsBLAST tool in the NCBI BLAST package to search the conserved domain database (35). The highly encoded secreted protein families having more than 50 members in the whole database are listed in Table 2. Preliminary functional analysis revealed that the fungal secretomes largely consist of enzymes, particularly hydrolases, which are used to breakdown carbohydrates, lipids, proteins and all other types of organic materials by fungi (Table 2). Furthermore, a total of 10 397 secreted proteins have GO annotations in UniProtKB. Among them, molecular functional classification using GOSlimViewer (http://agbase.msstate.edu/cgi-bin/tools/goslimviewer_select.pl) showed 43% were hydrolases including peptidases (Figure 3) (36). These enzymes have potential applications in biofuel production. The database user interface features an easy to use option to download predicted secretomes from completely sequenced fungal species. This provides a resource for further detailed species specific or interspecies comparative analysis.
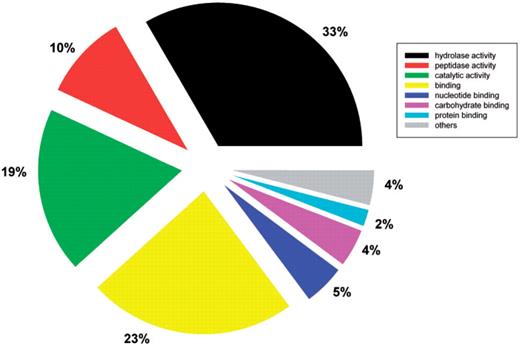
Molecular functional classification of fungal secreted proteins using GOSlimViewer.
| CDD functional domains . | Numbers . |
|---|---|
| pfam00135, COesterase, Carboxylesterase | 314 |
| pfam03443, Glyco hydro 61, Glycosyl hydrolase family 61 | 301 |
| COG0277, GlcD, FAD/FMN-containing dehydrogenases | 287 |
| cd04077, Peptidases S8 PCSK9 ProteinaseK like: Peptidase S8 family domain in ProteinaseK-like proteins | 223 |
| pfam00450, Peptidase S10, Serine carboxypeptidase | 215 |
| pfam00295, Glyco hydro 28, Glycosyl hydrolases family 28 | 207 |
| pfam00067, p450, Cytochrome P450 | 160 |
| pfam00933, Glyco hydro 3, Glycosyl hydrolase family 3 N terminal domain | 156 |
| cd05474, pepsin-like proteinases secreted from pathogens to degrade host proteins | 154 |
| COG2303, BetA, Choline dehydrogenase and related flavoproteins | 152 |
| pfam01083, Cutinase | 139 |
| pfam09362, DUF1996, Domain of unknown function (DUF1996) | 136 |
| pfam00264, Tyrosinase, Common central domain of tyrosinase | 130 |
| TIGR03388, ascorbase, L-ascorbate oxidase, plant type | 128 |
| cd04056, Peptidases S53, Peptidase domain in the S53 family | 124 |
| pfam04389, Peptidase M28, Peptidase family M28 | 122 |
| COG5309, COG5309, Exo-beta-1,3-glucanase | 121 |
| pfam04616, Glyco hydro 43, Glycosyl hydrolases family 43 | 114 |
| cd00519, Lipase 3, Lipase (class 3) | 106 |
| PRK02106, PRK02106, choline dehydrogenase | 100 |
| COG2730, BglC, Endoglucanase | 99 |
| pfam00328, Acid phosphat A, Histidine acid phosphatase | 98 |
| pfam03856, SUN, Beta-glucosidase (SUN family) | 97 |
| pfam07519, Tannase, Tannase and feruloyl esterase | 97 |
| smart00656, Amb all, Amb all domain | 94 |
| pfam00457, Glyco hydro 11, Glycosyl hydrolases family 11 | 92 |
| cd06097, Aspergillopepsin like: Aspergillopepsin like, aspartic proteases of fungal origin | 91 |
| cd02877, GH18 hevamine XipI class III | 88 |
| pfam00331, Glyco hydro 10, Glycosyl hydrolase family 10 | 88 |
| pfam01565, FAD binding 4, FAD binding domain | 87 |
| pfam03583, LIP, Secretory lipase | 87 |
| pfam03659, Glyco hydro 71, Glycosyl hydrolase family 71 | 87 |
| pfam01185, Hydrophobin, Fungal hydrophobin | 85 |
| pfam01532, Glyco hydro 47, Glycosyl hydrolase family 47 | 79 |
| cd02181, GH16 MLG1 glucanase | 78 |
| cd05471, Pepsin-like aspartic proteases, bilobal enzymes that cleave bonds in peptides at acidic pH | 77 |
| cd05384, SCP PRY1 like, SCP-like extracellular protein domain, PRY1-like sub-family restricted to fungi | 75 |
| cd07203, Fungal Phospholipase B-like; cPLA2 GrpIVA homologs; catalytic domain | 71 |
| pfam00840, Glyco hydro 7, Glycosyl hydrolase family 7 | 71 |
| pfam00150, Cellulase, Cellulase (glycosyl hydrolase family 5) | 70 |
| pfam11790, Glyco hydro cc, Glycosyl hydrolase catalytic core | 70 |
| pfam01522, Polysacc deac 1, Polysaccharide deacetylase | 69 |
| pfam07971, Glyco hydro 92, Glycosyl hydrolase family 92 | 68 |
| smart00636, Glyco 18, Glycosyl hydrolase family 18 | 68 |
| cd00842, MPP ASMase, acid sphingomyelinase and related proteins | 67 |
| cd03457, intradiol dioxygenase like, Intradiol dioxygenase supgroup | 67 |
| pfam03663, Glyco hydro 76, Glycosyl hydrolase family 76 | 67 |
| pfam05577, Peptidase S28, Serine carboxypeptidase S28 | 67 |
| pfam12296, HsbA, Hydrophobic surface binding protein A | 65 |
| cd02183, GH16 GPI glucanosyltransferase | 64 |
| COG0654, 2-polyprenyl-6-methoxyphenol hydroxylase and related FAD-dependent oxidoreductases | 63 |
| pfam01055, Glyco hydro 31, Glycosyl hydrolases family 31 | 62 |
| cd06248, Peptidase M14 Carboxypeptidase A/B-like subfamily | 61 |
| pfam02128, Peptidase M36, Fungalysin metallopeptidase (M36) | 61 |
| pfam04185, Phosphoesterase, Phosphoesterase family | 61 |
| pfam11765, Hyphal reg CWP, Hyphally regulated cell wall protein | 60 |
| pfam01328, Peroxidase 2, Peroxidase, family 2 | 59 |
| pfam01828, Peptidase A4, Peptidase A4 family | 58 |
| pfam03198, Glyco hydro 72, Glycolipid anchored surface protein | 57 |
| cd01846, Fatty acyltransferase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases | 56 |
| pfam02102, Peptidase M35, Deuterolysin metalloprotease (M35) | 56 |
| pfam00723, Glyco hydro 15, Glycosyl hydrolases family 15 | 54 |
| pfam00128, Alpha-amylase, Alpha amylase, catalytic domain | 53 |
| cd08588, Catalytic domain of Arabidopsis thaliana PI-PLC X domain-containing protein | 52 |
| PHA03247, PHA03247, large tegument protein UL36; Provisional | 52 |
| pfam01301, Glyco hydro 35, Glycosyl hydrolases family 35 | 51 |
| pfam11937, DUF3455, Protein of unknown function (DUF3455) | 51 |
| CDD functional domains . | Numbers . |
|---|---|
| pfam00135, COesterase, Carboxylesterase | 314 |
| pfam03443, Glyco hydro 61, Glycosyl hydrolase family 61 | 301 |
| COG0277, GlcD, FAD/FMN-containing dehydrogenases | 287 |
| cd04077, Peptidases S8 PCSK9 ProteinaseK like: Peptidase S8 family domain in ProteinaseK-like proteins | 223 |
| pfam00450, Peptidase S10, Serine carboxypeptidase | 215 |
| pfam00295, Glyco hydro 28, Glycosyl hydrolases family 28 | 207 |
| pfam00067, p450, Cytochrome P450 | 160 |
| pfam00933, Glyco hydro 3, Glycosyl hydrolase family 3 N terminal domain | 156 |
| cd05474, pepsin-like proteinases secreted from pathogens to degrade host proteins | 154 |
| COG2303, BetA, Choline dehydrogenase and related flavoproteins | 152 |
| pfam01083, Cutinase | 139 |
| pfam09362, DUF1996, Domain of unknown function (DUF1996) | 136 |
| pfam00264, Tyrosinase, Common central domain of tyrosinase | 130 |
| TIGR03388, ascorbase, L-ascorbate oxidase, plant type | 128 |
| cd04056, Peptidases S53, Peptidase domain in the S53 family | 124 |
| pfam04389, Peptidase M28, Peptidase family M28 | 122 |
| COG5309, COG5309, Exo-beta-1,3-glucanase | 121 |
| pfam04616, Glyco hydro 43, Glycosyl hydrolases family 43 | 114 |
| cd00519, Lipase 3, Lipase (class 3) | 106 |
| PRK02106, PRK02106, choline dehydrogenase | 100 |
| COG2730, BglC, Endoglucanase | 99 |
| pfam00328, Acid phosphat A, Histidine acid phosphatase | 98 |
| pfam03856, SUN, Beta-glucosidase (SUN family) | 97 |
| pfam07519, Tannase, Tannase and feruloyl esterase | 97 |
| smart00656, Amb all, Amb all domain | 94 |
| pfam00457, Glyco hydro 11, Glycosyl hydrolases family 11 | 92 |
| cd06097, Aspergillopepsin like: Aspergillopepsin like, aspartic proteases of fungal origin | 91 |
| cd02877, GH18 hevamine XipI class III | 88 |
| pfam00331, Glyco hydro 10, Glycosyl hydrolase family 10 | 88 |
| pfam01565, FAD binding 4, FAD binding domain | 87 |
| pfam03583, LIP, Secretory lipase | 87 |
| pfam03659, Glyco hydro 71, Glycosyl hydrolase family 71 | 87 |
| pfam01185, Hydrophobin, Fungal hydrophobin | 85 |
| pfam01532, Glyco hydro 47, Glycosyl hydrolase family 47 | 79 |
| cd02181, GH16 MLG1 glucanase | 78 |
| cd05471, Pepsin-like aspartic proteases, bilobal enzymes that cleave bonds in peptides at acidic pH | 77 |
| cd05384, SCP PRY1 like, SCP-like extracellular protein domain, PRY1-like sub-family restricted to fungi | 75 |
| cd07203, Fungal Phospholipase B-like; cPLA2 GrpIVA homologs; catalytic domain | 71 |
| pfam00840, Glyco hydro 7, Glycosyl hydrolase family 7 | 71 |
| pfam00150, Cellulase, Cellulase (glycosyl hydrolase family 5) | 70 |
| pfam11790, Glyco hydro cc, Glycosyl hydrolase catalytic core | 70 |
| pfam01522, Polysacc deac 1, Polysaccharide deacetylase | 69 |
| pfam07971, Glyco hydro 92, Glycosyl hydrolase family 92 | 68 |
| smart00636, Glyco 18, Glycosyl hydrolase family 18 | 68 |
| cd00842, MPP ASMase, acid sphingomyelinase and related proteins | 67 |
| cd03457, intradiol dioxygenase like, Intradiol dioxygenase supgroup | 67 |
| pfam03663, Glyco hydro 76, Glycosyl hydrolase family 76 | 67 |
| pfam05577, Peptidase S28, Serine carboxypeptidase S28 | 67 |
| pfam12296, HsbA, Hydrophobic surface binding protein A | 65 |
| cd02183, GH16 GPI glucanosyltransferase | 64 |
| COG0654, 2-polyprenyl-6-methoxyphenol hydroxylase and related FAD-dependent oxidoreductases | 63 |
| pfam01055, Glyco hydro 31, Glycosyl hydrolases family 31 | 62 |
| cd06248, Peptidase M14 Carboxypeptidase A/B-like subfamily | 61 |
| pfam02128, Peptidase M36, Fungalysin metallopeptidase (M36) | 61 |
| pfam04185, Phosphoesterase, Phosphoesterase family | 61 |
| pfam11765, Hyphal reg CWP, Hyphally regulated cell wall protein | 60 |
| pfam01328, Peroxidase 2, Peroxidase, family 2 | 59 |
| pfam01828, Peptidase A4, Peptidase A4 family | 58 |
| pfam03198, Glyco hydro 72, Glycolipid anchored surface protein | 57 |
| cd01846, Fatty acyltransferase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases | 56 |
| pfam02102, Peptidase M35, Deuterolysin metalloprotease (M35) | 56 |
| pfam00723, Glyco hydro 15, Glycosyl hydrolases family 15 | 54 |
| pfam00128, Alpha-amylase, Alpha amylase, catalytic domain | 53 |
| cd08588, Catalytic domain of Arabidopsis thaliana PI-PLC X domain-containing protein | 52 |
| PHA03247, PHA03247, large tegument protein UL36; Provisional | 52 |
| pfam01301, Glyco hydro 35, Glycosyl hydrolases family 35 | 51 |
| pfam11937, DUF3455, Protein of unknown function (DUF3455) | 51 |
| CDD functional domains . | Numbers . |
|---|---|
| pfam00135, COesterase, Carboxylesterase | 314 |
| pfam03443, Glyco hydro 61, Glycosyl hydrolase family 61 | 301 |
| COG0277, GlcD, FAD/FMN-containing dehydrogenases | 287 |
| cd04077, Peptidases S8 PCSK9 ProteinaseK like: Peptidase S8 family domain in ProteinaseK-like proteins | 223 |
| pfam00450, Peptidase S10, Serine carboxypeptidase | 215 |
| pfam00295, Glyco hydro 28, Glycosyl hydrolases family 28 | 207 |
| pfam00067, p450, Cytochrome P450 | 160 |
| pfam00933, Glyco hydro 3, Glycosyl hydrolase family 3 N terminal domain | 156 |
| cd05474, pepsin-like proteinases secreted from pathogens to degrade host proteins | 154 |
| COG2303, BetA, Choline dehydrogenase and related flavoproteins | 152 |
| pfam01083, Cutinase | 139 |
| pfam09362, DUF1996, Domain of unknown function (DUF1996) | 136 |
| pfam00264, Tyrosinase, Common central domain of tyrosinase | 130 |
| TIGR03388, ascorbase, L-ascorbate oxidase, plant type | 128 |
| cd04056, Peptidases S53, Peptidase domain in the S53 family | 124 |
| pfam04389, Peptidase M28, Peptidase family M28 | 122 |
| COG5309, COG5309, Exo-beta-1,3-glucanase | 121 |
| pfam04616, Glyco hydro 43, Glycosyl hydrolases family 43 | 114 |
| cd00519, Lipase 3, Lipase (class 3) | 106 |
| PRK02106, PRK02106, choline dehydrogenase | 100 |
| COG2730, BglC, Endoglucanase | 99 |
| pfam00328, Acid phosphat A, Histidine acid phosphatase | 98 |
| pfam03856, SUN, Beta-glucosidase (SUN family) | 97 |
| pfam07519, Tannase, Tannase and feruloyl esterase | 97 |
| smart00656, Amb all, Amb all domain | 94 |
| pfam00457, Glyco hydro 11, Glycosyl hydrolases family 11 | 92 |
| cd06097, Aspergillopepsin like: Aspergillopepsin like, aspartic proteases of fungal origin | 91 |
| cd02877, GH18 hevamine XipI class III | 88 |
| pfam00331, Glyco hydro 10, Glycosyl hydrolase family 10 | 88 |
| pfam01565, FAD binding 4, FAD binding domain | 87 |
| pfam03583, LIP, Secretory lipase | 87 |
| pfam03659, Glyco hydro 71, Glycosyl hydrolase family 71 | 87 |
| pfam01185, Hydrophobin, Fungal hydrophobin | 85 |
| pfam01532, Glyco hydro 47, Glycosyl hydrolase family 47 | 79 |
| cd02181, GH16 MLG1 glucanase | 78 |
| cd05471, Pepsin-like aspartic proteases, bilobal enzymes that cleave bonds in peptides at acidic pH | 77 |
| cd05384, SCP PRY1 like, SCP-like extracellular protein domain, PRY1-like sub-family restricted to fungi | 75 |
| cd07203, Fungal Phospholipase B-like; cPLA2 GrpIVA homologs; catalytic domain | 71 |
| pfam00840, Glyco hydro 7, Glycosyl hydrolase family 7 | 71 |
| pfam00150, Cellulase, Cellulase (glycosyl hydrolase family 5) | 70 |
| pfam11790, Glyco hydro cc, Glycosyl hydrolase catalytic core | 70 |
| pfam01522, Polysacc deac 1, Polysaccharide deacetylase | 69 |
| pfam07971, Glyco hydro 92, Glycosyl hydrolase family 92 | 68 |
| smart00636, Glyco 18, Glycosyl hydrolase family 18 | 68 |
| cd00842, MPP ASMase, acid sphingomyelinase and related proteins | 67 |
| cd03457, intradiol dioxygenase like, Intradiol dioxygenase supgroup | 67 |
| pfam03663, Glyco hydro 76, Glycosyl hydrolase family 76 | 67 |
| pfam05577, Peptidase S28, Serine carboxypeptidase S28 | 67 |
| pfam12296, HsbA, Hydrophobic surface binding protein A | 65 |
| cd02183, GH16 GPI glucanosyltransferase | 64 |
| COG0654, 2-polyprenyl-6-methoxyphenol hydroxylase and related FAD-dependent oxidoreductases | 63 |
| pfam01055, Glyco hydro 31, Glycosyl hydrolases family 31 | 62 |
| cd06248, Peptidase M14 Carboxypeptidase A/B-like subfamily | 61 |
| pfam02128, Peptidase M36, Fungalysin metallopeptidase (M36) | 61 |
| pfam04185, Phosphoesterase, Phosphoesterase family | 61 |
| pfam11765, Hyphal reg CWP, Hyphally regulated cell wall protein | 60 |
| pfam01328, Peroxidase 2, Peroxidase, family 2 | 59 |
| pfam01828, Peptidase A4, Peptidase A4 family | 58 |
| pfam03198, Glyco hydro 72, Glycolipid anchored surface protein | 57 |
| cd01846, Fatty acyltransferase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases | 56 |
| pfam02102, Peptidase M35, Deuterolysin metalloprotease (M35) | 56 |
| pfam00723, Glyco hydro 15, Glycosyl hydrolases family 15 | 54 |
| pfam00128, Alpha-amylase, Alpha amylase, catalytic domain | 53 |
| cd08588, Catalytic domain of Arabidopsis thaliana PI-PLC X domain-containing protein | 52 |
| PHA03247, PHA03247, large tegument protein UL36; Provisional | 52 |
| pfam01301, Glyco hydro 35, Glycosyl hydrolases family 35 | 51 |
| pfam11937, DUF3455, Protein of unknown function (DUF3455) | 51 |
| CDD functional domains . | Numbers . |
|---|---|
| pfam00135, COesterase, Carboxylesterase | 314 |
| pfam03443, Glyco hydro 61, Glycosyl hydrolase family 61 | 301 |
| COG0277, GlcD, FAD/FMN-containing dehydrogenases | 287 |
| cd04077, Peptidases S8 PCSK9 ProteinaseK like: Peptidase S8 family domain in ProteinaseK-like proteins | 223 |
| pfam00450, Peptidase S10, Serine carboxypeptidase | 215 |
| pfam00295, Glyco hydro 28, Glycosyl hydrolases family 28 | 207 |
| pfam00067, p450, Cytochrome P450 | 160 |
| pfam00933, Glyco hydro 3, Glycosyl hydrolase family 3 N terminal domain | 156 |
| cd05474, pepsin-like proteinases secreted from pathogens to degrade host proteins | 154 |
| COG2303, BetA, Choline dehydrogenase and related flavoproteins | 152 |
| pfam01083, Cutinase | 139 |
| pfam09362, DUF1996, Domain of unknown function (DUF1996) | 136 |
| pfam00264, Tyrosinase, Common central domain of tyrosinase | 130 |
| TIGR03388, ascorbase, L-ascorbate oxidase, plant type | 128 |
| cd04056, Peptidases S53, Peptidase domain in the S53 family | 124 |
| pfam04389, Peptidase M28, Peptidase family M28 | 122 |
| COG5309, COG5309, Exo-beta-1,3-glucanase | 121 |
| pfam04616, Glyco hydro 43, Glycosyl hydrolases family 43 | 114 |
| cd00519, Lipase 3, Lipase (class 3) | 106 |
| PRK02106, PRK02106, choline dehydrogenase | 100 |
| COG2730, BglC, Endoglucanase | 99 |
| pfam00328, Acid phosphat A, Histidine acid phosphatase | 98 |
| pfam03856, SUN, Beta-glucosidase (SUN family) | 97 |
| pfam07519, Tannase, Tannase and feruloyl esterase | 97 |
| smart00656, Amb all, Amb all domain | 94 |
| pfam00457, Glyco hydro 11, Glycosyl hydrolases family 11 | 92 |
| cd06097, Aspergillopepsin like: Aspergillopepsin like, aspartic proteases of fungal origin | 91 |
| cd02877, GH18 hevamine XipI class III | 88 |
| pfam00331, Glyco hydro 10, Glycosyl hydrolase family 10 | 88 |
| pfam01565, FAD binding 4, FAD binding domain | 87 |
| pfam03583, LIP, Secretory lipase | 87 |
| pfam03659, Glyco hydro 71, Glycosyl hydrolase family 71 | 87 |
| pfam01185, Hydrophobin, Fungal hydrophobin | 85 |
| pfam01532, Glyco hydro 47, Glycosyl hydrolase family 47 | 79 |
| cd02181, GH16 MLG1 glucanase | 78 |
| cd05471, Pepsin-like aspartic proteases, bilobal enzymes that cleave bonds in peptides at acidic pH | 77 |
| cd05384, SCP PRY1 like, SCP-like extracellular protein domain, PRY1-like sub-family restricted to fungi | 75 |
| cd07203, Fungal Phospholipase B-like; cPLA2 GrpIVA homologs; catalytic domain | 71 |
| pfam00840, Glyco hydro 7, Glycosyl hydrolase family 7 | 71 |
| pfam00150, Cellulase, Cellulase (glycosyl hydrolase family 5) | 70 |
| pfam11790, Glyco hydro cc, Glycosyl hydrolase catalytic core | 70 |
| pfam01522, Polysacc deac 1, Polysaccharide deacetylase | 69 |
| pfam07971, Glyco hydro 92, Glycosyl hydrolase family 92 | 68 |
| smart00636, Glyco 18, Glycosyl hydrolase family 18 | 68 |
| cd00842, MPP ASMase, acid sphingomyelinase and related proteins | 67 |
| cd03457, intradiol dioxygenase like, Intradiol dioxygenase supgroup | 67 |
| pfam03663, Glyco hydro 76, Glycosyl hydrolase family 76 | 67 |
| pfam05577, Peptidase S28, Serine carboxypeptidase S28 | 67 |
| pfam12296, HsbA, Hydrophobic surface binding protein A | 65 |
| cd02183, GH16 GPI glucanosyltransferase | 64 |
| COG0654, 2-polyprenyl-6-methoxyphenol hydroxylase and related FAD-dependent oxidoreductases | 63 |
| pfam01055, Glyco hydro 31, Glycosyl hydrolases family 31 | 62 |
| cd06248, Peptidase M14 Carboxypeptidase A/B-like subfamily | 61 |
| pfam02128, Peptidase M36, Fungalysin metallopeptidase (M36) | 61 |
| pfam04185, Phosphoesterase, Phosphoesterase family | 61 |
| pfam11765, Hyphal reg CWP, Hyphally regulated cell wall protein | 60 |
| pfam01328, Peroxidase 2, Peroxidase, family 2 | 59 |
| pfam01828, Peptidase A4, Peptidase A4 family | 58 |
| pfam03198, Glyco hydro 72, Glycolipid anchored surface protein | 57 |
| cd01846, Fatty acyltransferase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases | 56 |
| pfam02102, Peptidase M35, Deuterolysin metalloprotease (M35) | 56 |
| pfam00723, Glyco hydro 15, Glycosyl hydrolases family 15 | 54 |
| pfam00128, Alpha-amylase, Alpha amylase, catalytic domain | 53 |
| cd08588, Catalytic domain of Arabidopsis thaliana PI-PLC X domain-containing protein | 52 |
| PHA03247, PHA03247, large tegument protein UL36; Provisional | 52 |
| pfam01301, Glyco hydro 35, Glycosyl hydrolases family 35 | 51 |
| pfam11937, DUF3455, Protein of unknown function (DUF3455) | 51 |
Discussion
While constructing our database, a similar fungal secretome database (FSD, http://fsd.snu.ac.kr/) was published by Choi et al. (37). However, there are several important differences between the two databases (Table 3). We used RefSeq data while the FSD used only completely sequenced fungal genome data including some ‘work in progress’ genomes (37). The prediction methods used for identification of secreted proteins were also different. The FSD used a three-layer hierarchical identification rule based on 9 different programs and considered entries to be secreted proteins as long as any one of the tools predicted it to be secreted, thus the number of secreted proteins were much higher than the number predicted in our database. For example, in A. niger, we predicted 832 secreted proteins in the strain CBS 513.88, while Choi et al. (37) predicted 1831 secreted proteins in the same strain and 2616 secreted proteins in the ATCC1015 strain in the FSD (37). However, there were only from 691 to 881 proteins which were predicted to be secreted, with 160 of them being confirmed experimentally in the ATCC1015 strain by Tsang et al. (8). Thus, we believe the methods used in the FSD significantly over-estimated the number of secreted proteins in fungi. In addition, the search for the FSD is limited to using the sequence locus name and can not be searched with NCBI gi and accession number, UniProt accession number or keywords. There is also not a curation tool available for the community annotation in FSD (37).
| . | FSD . | FunSecKB . |
|---|---|---|
| Data source | Fungal genomes | Fungal proteins in RefSeq |
| Prediction tools | SignalP3.0; SigCleave; SigPred; RPSP; TMHMM2.0c; TargetP1.1b; PsortII; PredictNLS; SecretomeP1.0f | SignalP 3.0; Phobius1.01; WolfPsort0.2; TargetP1.1b, TMHMM2.0c; PS-Scan |
| Data access | Sequence locus name; BLAST | Keywords, RefSeq gi or accession, UniProt accession; BLAST |
| Community curation tool | Not available | Available |
| . | FSD . | FunSecKB . |
|---|---|---|
| Data source | Fungal genomes | Fungal proteins in RefSeq |
| Prediction tools | SignalP3.0; SigCleave; SigPred; RPSP; TMHMM2.0c; TargetP1.1b; PsortII; PredictNLS; SecretomeP1.0f | SignalP 3.0; Phobius1.01; WolfPsort0.2; TargetP1.1b, TMHMM2.0c; PS-Scan |
| Data access | Sequence locus name; BLAST | Keywords, RefSeq gi or accession, UniProt accession; BLAST |
| Community curation tool | Not available | Available |
| . | FSD . | FunSecKB . |
|---|---|---|
| Data source | Fungal genomes | Fungal proteins in RefSeq |
| Prediction tools | SignalP3.0; SigCleave; SigPred; RPSP; TMHMM2.0c; TargetP1.1b; PsortII; PredictNLS; SecretomeP1.0f | SignalP 3.0; Phobius1.01; WolfPsort0.2; TargetP1.1b, TMHMM2.0c; PS-Scan |
| Data access | Sequence locus name; BLAST | Keywords, RefSeq gi or accession, UniProt accession; BLAST |
| Community curation tool | Not available | Available |
| . | FSD . | FunSecKB . |
|---|---|---|
| Data source | Fungal genomes | Fungal proteins in RefSeq |
| Prediction tools | SignalP3.0; SigCleave; SigPred; RPSP; TMHMM2.0c; TargetP1.1b; PsortII; PredictNLS; SecretomeP1.0f | SignalP 3.0; Phobius1.01; WolfPsort0.2; TargetP1.1b, TMHMM2.0c; PS-Scan |
| Data access | Sequence locus name; BLAST | Keywords, RefSeq gi or accession, UniProt accession; BLAST |
| Community curation tool | Not available | Available |
In addition to the signal-peptide dependent secreted proteins using the classical ER-Golgi secretory pathway, there are non-classical, signal peptide independent, secretory pathways in all domains of organisms. Mammalian and bacterial leadless secreted proteins have been collected and used to implement the prediction software, SecretomeP, for predicting these proteins (http://www.cbs.dtu.dk/services/SecretomeP/) (38,39). The tool has not been trained with fungal-specific data and the accuracy for predicting fungal non-classical secreted protein could not be evaluated, thus we did not include this tool in our data processing. Although the FSD used SecretomeP to predict non-classical secreted proteins, the predicted secreted proteins were not included in the secretome analysis; including them would make the putative secretome >40% of whole proteome (37). Nevertheless, the FunSecKB and the FSD databases could complement each other as different data sources, prediction tools and data access utilities were implemented.
In summary, we constructed FunSecKB to identify, annotate and curate the secreted proteins in fungi. The data can be searched using protein identifiers or keywords, and by species. Most of the secreted proteins are currently predicted by computational tools. However, the community can use the curation module implemented in our site to manually curate subcellular locations of fungal proteins having experimental evidence. The resource described in the work is expected to provide a query and curation system that will help the community to further understand the secretome biology and explore various potential applications of fungal secreted proteins in bio-processing or environmental remediation industries.
Acknowledgements
We thank Gary Walker at YSU and the anonymous reviewers for providing helpful comments on improving the article.
Funding
Youngstown State University (YSU) Research Council grant (2009-2010 #04-10 to X.J.M.); YSU research professorship (to X.J.M.); College of Science, Technology, Engineering, and Mathematics Dean’s reassigned time (to X.J.M.). Funding for open access charge: the School of Graduate Studies and Research, Youngstown State University, Ohio, USA.
Conflict of interest. None Declared.



