-
PDF
- Split View
-
Views
-
Cite
Cite
Asma N Cheema, Samantha L Rosenthal, M Ilyas Kamboh, Proficiency of data interpretation: identification of signaling SNPs/specific loci for coronary artery disease, Database, Volume 2017, 2017, bax078, https://doi.org/10.1093/database/bax078
Close - Share Icon Share
Abstract
Coronary artery disease (CAD) is a complex disorder involving both genetic and non-genetic factors. Genome-wide association studies (GWAS) have identified hundreds of single nucleotides polymorphisms (SNPs) tagging over > 40 CAD risk loci. We hypothesized that some non-coding variants might directly regulate the gene expression rather than tagging a nearby locus. We used RegulomeDB to examine regulatory functions of 58 SNPs identified in two GWAS and those SNPs in linkage disequilibrium (LD) (r2 ≥ 0.80) with the GWAS SNPs. Of the tested 1200 SNPs, 858 returned scores of 1–6 by RegulomeDB. Of these 858 SNPs, 97 were predicted to have regulatory functions with RegulomeDB score of < 3. Notably, only 8 of the 97 predicted regulatory variants were genome-wide significant SNPs (LIPA/rs2246833, RegulomeDB score = 1b; ZC3HC1/rs11556924, CYP17A1-CNNM2-NT5C2/rs12413409, APOE-APOC1/rs2075650 and UBE2Z/rs46522, each with a RegulomeDB score = 1f; ZNF259-APOA5-APOA1/rs964184, SMG6/rs2281727 and COL4A1-COL4A2/rs4773144, each with a RegulomeDB score = 2b). The remainder 89 functional SNPs were in linkage disequilibrium with GWAS SNPs. This study supports the hypothesis that some of the non-coding variants are true signals via regulation of gene expression at transcription level. Our study indicates that RegulomeDB is a useful database to examine the putative functions of large number of genetic variants and it may help to identify a true association among multiple tagged SNPs in a complex disease, such as CAD.
Background
Most human DNA sequence is non-coding (98%) and hence only small portion (2%) of human genome encodes proteins (1). Although the pathogenesis of monogenic disorders is largely explained, it has been difficult to determine the underlying mechanisms of complex disorders like coronary artery disease (CAD). Before the development of genome-wide association studies (GWAS), only the APOE*4 allele showed consistent association with the risk of CAD across many populations (2–5).
The hypothesis-free GWAS approach was designed with the assumption that common DNA variants explain the bulk of the variation in common diseases (6). About 90% of GWAS-implicated variants, exert only minimal to modest effect sizes on disease phenotypes, and they are present in non-coding rather than coding regions (7). Highly sensitive molecular and computational techniques have identified different regulatory elements (DNAse hypersensitive regions, sequences affecting the binding of transcription factors and promoters or enhancers) in intergenic regions (8). Common variants located in one of these regulatory elements may affect gene expression. To predict the role of these variants in gene regulation and to differentiate between physically tagged and functional single nucleotides polymorphism (SNPs), many databases have been created (9). RegulomeDB is one of such databases that describes the role of these variants in transcriptional regulation.
Similar to many other complex diseases, GWAS have identified hundreds of risk variants associated with CAD that need to be analyzed for their functional role in gene expression (10). Recently, we have used SNAP Webportal and Regulome DB to identify potential regulatory function of variants in associated risk loci for Alzheimer’s disease (11). In this study, we have applied the same approach to identify the regulatory nature of GWAS-implicated variants with CAD and those that are in linkage disequilibrium (LD) with these variants.
Objective
The objective of our study was to assess the GWAS-implicated CAD variants and those variants in LD with GWAS variants for their potential regulatory effects on gene transcription using bioinformatics tools.
Materials and methods
SNPs selection
A total of 58 SNPs within 54 CAD loci was selected, including 52 with accepted genome-wide significant threshold (P < 5 × 10−8) and 6 with suggestive associations (P > 5 × 10−8) identified in two GWAS (12, 13). Detailed information on the selected 58 SNPs is provided in Supplementary Table S1.
Linkage disequilibrium
For the LD assessment of the selected 58 SNPs, we used SNAP web portal (https://www.broadinstitute.org/mpg/snap/, accessed 13 July 2016) (14) (Supplementary Table S2). SNAP contains data from the Northern European from Utah (CEU) population derived from the 1000 Genomes Pilot Project 1 and three different releases of the International-Hap Map Project. We used data from both the 1000 Genomes Project and HapMap 3 (release 2) to identify SNPs in strong LD (r2 ≥ 0.80) with our SNPs of interest. We did not select an array bound search, and query SNPs were included in the output. We performed the search at three thresholds—r2 ≥ 0.80, r2 ≥ 0.90 and r2 ≥ 1.0—for both SNP datasets and identified a total of 1,200 SNPs in LD with the 58 published GWAS SNPs, including the GWAS SNPs themselves. As shown in Table 1, the number of proxy SNPs decreased with the increased level of r2.
Number of SNPs in LD for all published GWAS SNPs for HapMap3 and 1000 genomes populations at tested r2 threshold
| . | . | LD . | . |
|---|---|---|---|
| r2threshold | 0.80 | 0.90 | 1.0 |
| 1000 Genomes | 1176 | 928 | 480 |
| Hap Map3 | 210 | 157 | 74 |
| Total (overlaps removed) | 1200 | 934 | 485 |
| . | . | LD . | . |
|---|---|---|---|
| r2threshold | 0.80 | 0.90 | 1.0 |
| 1000 Genomes | 1176 | 928 | 480 |
| Hap Map3 | 210 | 157 | 74 |
| Total (overlaps removed) | 1200 | 934 | 485 |
Number of SNPs in LD for all published GWAS SNPs for HapMap3 and 1000 genomes populations at tested r2 threshold
| . | . | LD . | . |
|---|---|---|---|
| r2threshold | 0.80 | 0.90 | 1.0 |
| 1000 Genomes | 1176 | 928 | 480 |
| Hap Map3 | 210 | 157 | 74 |
| Total (overlaps removed) | 1200 | 934 | 485 |
| . | . | LD . | . |
|---|---|---|---|
| r2threshold | 0.80 | 0.90 | 1.0 |
| 1000 Genomes | 1176 | 928 | 480 |
| Hap Map3 | 210 | 157 | 74 |
| Total (overlaps removed) | 1200 | 934 | 485 |
Functional assessment of CAD-associated SNPs
We used RegulomeDB to identify potentially functional SNPs among the 1200 SNPs of interest. Regulome DB is a database that scores SNPs functionality based upon experimental data, such as its existence in a DNAase hypersensitive site or transcription factor binding site. These regions have been characterized biochemically, and data are drawn from published literature, Gene Expression Omnibus and ENCODE project that include a total of 962 experimental datasets, covering over 100 tissues and cell lines and representing nearly 60 million annotations. The output data can be mapped to Human genome version 19. It is a user friendly and freely accessible database (http://www.regulomedb.org/accessed 17 July 2016) (15). The functional Grades 1–6 of RegulomeDB are given in Table 2. SNPs showing the strongest evidence of being regulatory (affecting the binding of transcription factor) are given a score of 1 and SNPs demonstrating the least evidence of being functional are given a score of 6.
| Category . | Description . |
|---|---|
| Likely to affect binding and linked to expression of a gene target | |
| 1b | eQTL + TF binding + any motif + DNase footprint + DNase peak |
| 1c | eQTL + TF binding + matched TF motif + DNase peak |
| 1d | eQTL + TF binding + any motif + DNase peak |
| 1e | eQTL + TF binding + matched TF motif |
| 1f | eQTL + TF binding/DNase peak |
| Likely to affect binding | |
| 2a | TF binding + matched TF motif + matched DNase footprint + DNase peak |
| 2b | TF binding + any motif + DNase footprint + DNase peak |
| 2c | TF binding + matched TF motif + DNase peak |
| Less likely to affect binding | |
| 3a | TF binding + any motif + DNase peak |
| 3b | TF binding + matched TF motif |
| Minimal binding evidence | |
| 4 | TF binding + DNase peak |
| 5 | TF binding or DNase peak |
| 6 | Motif hit |
| Category . | Description . |
|---|---|
| Likely to affect binding and linked to expression of a gene target | |
| 1b | eQTL + TF binding + any motif + DNase footprint + DNase peak |
| 1c | eQTL + TF binding + matched TF motif + DNase peak |
| 1d | eQTL + TF binding + any motif + DNase peak |
| 1e | eQTL + TF binding + matched TF motif |
| 1f | eQTL + TF binding/DNase peak |
| Likely to affect binding | |
| 2a | TF binding + matched TF motif + matched DNase footprint + DNase peak |
| 2b | TF binding + any motif + DNase footprint + DNase peak |
| 2c | TF binding + matched TF motif + DNase peak |
| Less likely to affect binding | |
| 3a | TF binding + any motif + DNase peak |
| 3b | TF binding + matched TF motif |
| Minimal binding evidence | |
| 4 | TF binding + DNase peak |
| 5 | TF binding or DNase peak |
| 6 | Motif hit |
| Category . | Description . |
|---|---|
| Likely to affect binding and linked to expression of a gene target | |
| 1b | eQTL + TF binding + any motif + DNase footprint + DNase peak |
| 1c | eQTL + TF binding + matched TF motif + DNase peak |
| 1d | eQTL + TF binding + any motif + DNase peak |
| 1e | eQTL + TF binding + matched TF motif |
| 1f | eQTL + TF binding/DNase peak |
| Likely to affect binding | |
| 2a | TF binding + matched TF motif + matched DNase footprint + DNase peak |
| 2b | TF binding + any motif + DNase footprint + DNase peak |
| 2c | TF binding + matched TF motif + DNase peak |
| Less likely to affect binding | |
| 3a | TF binding + any motif + DNase peak |
| 3b | TF binding + matched TF motif |
| Minimal binding evidence | |
| 4 | TF binding + DNase peak |
| 5 | TF binding or DNase peak |
| 6 | Motif hit |
| Category . | Description . |
|---|---|
| Likely to affect binding and linked to expression of a gene target | |
| 1b | eQTL + TF binding + any motif + DNase footprint + DNase peak |
| 1c | eQTL + TF binding + matched TF motif + DNase peak |
| 1d | eQTL + TF binding + any motif + DNase peak |
| 1e | eQTL + TF binding + matched TF motif |
| 1f | eQTL + TF binding/DNase peak |
| Likely to affect binding | |
| 2a | TF binding + matched TF motif + matched DNase footprint + DNase peak |
| 2b | TF binding + any motif + DNase footprint + DNase peak |
| 2c | TF binding + matched TF motif + DNase peak |
| Less likely to affect binding | |
| 3a | TF binding + any motif + DNase peak |
| 3b | TF binding + matched TF motif |
| Minimal binding evidence | |
| 4 | TF binding + DNase peak |
| 5 | TF binding or DNase peak |
| 6 | Motif hit |
Results
Among the 1200 SNPs evaluated with RegulomeDB, 342 had no data (Supplementary Table S3). Of the 858 SNPs for which RegulomeDB provided a score, 97 had a score of <3 (likely to affect the binding) and among these only 8 SNPs were genome-wide significant, including LIPA/rs2246833 (RegulomeDB score = 1b; eQTL in monocytes), ZC3HC1/rs11556924 (RegulomeDB score = 1f; eQTL in monocytes), CYP17A1-CNNM2-NT5C2/rs12413409 (RegulomeDB score = 1f; eQTL in monocytes and lymphoblasts), APOE-APOC1/rs2075650, and UBE2Z/rs46522 (RegulomeDB score = 1f; eQTL in monocytes), ZNF259-APOA5-APOA1/rs964184, UBE2Z/rs46522, SMG6/rs2281727, COL4A1-COL4A2/rs4773144 (RegulomeDB score =2b; eQTLs in monocytes and lymphoblasts). A flow chart summarizes these results (Figure 1) . The remaining 89 SNPs with RegulomeDB scores < 3 were not identified in GWAS but they were in LD (r2 ≥ 0.80) with the 29 GWAS reported SNPs. A summary of the regulatory SNPs in LD with GWAS SNPs is provided in Table 3.
Functional SNPs (RegluomeDB Score < 3) in LD (r2 ≥ 0.80) with published GWAS SNPs
| GWAS SNPs . | Functional proxy SNPs . | Regulome DB score . |
|---|---|---|
| LIPA/rs2246833 | LIPA/rs1332327 | 2b |
| LIPA/rs1332328 | 2b | |
| LIPA/rs1412444 | 1d | |
| LIPA/rs2246833a | 1b | |
| LIPA/rs2250644 | 2b | |
| ZC3HC1/rs11556924 | ZC3HC1/rs11556924a | 1f |
| CYP17A1-CNNM2-NT5C2/rs12413409 | AS3MT/rs11191454 | 1f |
| BORCS7-ASMT/rs4409766 | 1f | |
| CNNM2/rs10883808 | 1f | |
| MAT2A/rs1446668 | 2a | |
| NT5C2/rs10883832 | 1f | |
| CNNM2/rs11191479 | 1f | |
| NT5C2/rs11191557 | 1f | |
| CNNM2/rs11191499 | 1f | |
| NT5C2/rs11191558 | 1f | |
| CNNM2/rs11191514 | 1f | |
| NT5C2/rs11191580 | 1f | |
| CNNM2/rs11191515 | 1f | |
| NT5C2/rs11191582 | 1f | |
| CNNM2/rs12221064 | 2b | |
| NT5C2/rs12412038 | 1f | |
| CNNM2/rs12411886 | 1f | |
| NT5C2/rs12413046 | 1f | |
| CNNM2/rs12413409a | 1f | |
| NT5C2/rs9633712 | 1e | |
| CNNM2/rs17115213 | 1f | |
| NT5C2/rs11191548 | 1f | |
| CNNM2/rs2297787 | 2a | |
| CNNM2/rs3781285 | 1f | |
| CNNM2/rs943037 | 1f | |
| CNNM2/rs12219901 | 2b | |
| APOE-APOC1/TOMM40/rs2075650 | APOE-APOC1/rs2075650a | 1f |
| UBE2Z/rs46522 | GIP/rs2291725 | 1f |
| GIP/rs4794004 | 1d | |
| SNF8/rs1994970 | 1f | |
| SNF8/rs4793992 | 1f | |
| UBE2Z/rs12601672 | 2b | |
| UBE2Z/rs15563 | 1f | |
| UBE2Z/rs3744608 | 2a | |
| UBE2Z/rs3848460 | 1f | |
| UBE2Z/rs46522a | 1f | |
| UBE2Z/rs11079844 | 1f | |
| ZNF259-APOA5-APOA1/rs964184 | ZNF259-APOA5-APOA1/rs964184 | 1f |
| SMG6/rs2281727 | SMG6/rs2281727a | 2b |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| COL4A1-COL4A2/rs4773144 | COL4A1-COL4A2/rs4773144a | 2b |
| ABO/rs579459 | ABO/rs649129 | 2b |
| ADMTS7/rs7173743 | LOC105370915/rs5029904 | 2b |
| PHACTR1/rs4773143 | 2b | |
| CXCL12/rs501120 | CXCL12/rs518594 | 2b |
| CXCL12/rs1746052 | 2b | |
| FURIN-FES/rs17514846 | FES/rs1894401 | 1b |
| HHIPL1/rs2895811 | HHIPL1/rs28391527 | 2b |
| HHIPL1/rs4624107 | 2b | |
| HHIPL1/rs7145262 | 2b | |
| IL6R/rs4845625 | IL6R/rs7549250 | 2b |
| IL6R/rs7549338 | 2b | |
| IL6R/rs7553796 | 2b | |
| KCNE2/rs9982601 | KCNE2/rs28591415 | 2b |
| KIAA1462/rs2505083 | KIAA1462/rs3739998 | 2b |
| LPL/rs264 | LPL/rs271 | 1f |
| LPL/rs3779788 | 2b | |
| MIA3/rs17465637 | MIA3/rs17163301 | 2b |
| PLG/rs4252120 | PLG/rs4252126 | 1f |
| PLG/rs4252135 | 1f | |
| PPAP2B/rs17114036 | LOC101929929/rs72664304 | 2a |
| PLPP3/rs4634932 | 1f | |
| SLC22A4-SLC22A5/rs273909 | SLC22A5/rs17689550 | 1f |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| SORT1/rs602633 | CELSR2/rs12740374 | 2b |
| CELSR2/rs629301 | 1f | |
| CELSR2/rs646776 | 1f | |
| TRIB1/rs2954029 | LOC105375745/rs2980853 | 2b |
| LOC105375745/rs2001844 | 2b | |
| LOC105375745/rs6982636 | 2b | |
| TRIB1/rs2980856 | 2b | |
| VAMP5-VAMP8-GGCX/rs1561198 | GGCX/rs6738645 | 1f |
| GGCX/rs10187424 | 1f | |
| VAMP8/rs1009 | 1b | |
| GGCX/rs6547621 | 1f | |
| VAMP8/rs1348818 | 1f | |
| GGCX/rs2886722 | 1f | |
| VAMP8/rs3770098 | 1f | |
| VAMP8/rs6757263 | 1f | |
| WDR12/rs6725887 | ICA1L/rs72934715 | 2b |
| NBEAL1/rs2351524 | 1f | |
| WDR12/rs72936852 | 2b | |
| NBEAL1/rs4675310 | 1f | |
| NBEAL1/rs72934512 | 2b | |
| REST-NOA1/rs17087335 | REST/rs2227901 | 1f |
| REST-NOA1/rs7687767 | 1d | |
| SWAP70/rs10840293 | SWAP70/rs93138 | 1f |
| SWAP70/rs360136 | 1f | |
| SMAD3/rs56062135 | SMAD3/rs17293632 | 2a |
| SMAD3/rs1866316 | 2b | |
| MTERF1/rs8032739 | 2b | |
| CDKN2BAS1/rs1333049 | CDKN2BAS1/rs4977574 | 2c |
| GWAS SNPs . | Functional proxy SNPs . | Regulome DB score . |
|---|---|---|
| LIPA/rs2246833 | LIPA/rs1332327 | 2b |
| LIPA/rs1332328 | 2b | |
| LIPA/rs1412444 | 1d | |
| LIPA/rs2246833a | 1b | |
| LIPA/rs2250644 | 2b | |
| ZC3HC1/rs11556924 | ZC3HC1/rs11556924a | 1f |
| CYP17A1-CNNM2-NT5C2/rs12413409 | AS3MT/rs11191454 | 1f |
| BORCS7-ASMT/rs4409766 | 1f | |
| CNNM2/rs10883808 | 1f | |
| MAT2A/rs1446668 | 2a | |
| NT5C2/rs10883832 | 1f | |
| CNNM2/rs11191479 | 1f | |
| NT5C2/rs11191557 | 1f | |
| CNNM2/rs11191499 | 1f | |
| NT5C2/rs11191558 | 1f | |
| CNNM2/rs11191514 | 1f | |
| NT5C2/rs11191580 | 1f | |
| CNNM2/rs11191515 | 1f | |
| NT5C2/rs11191582 | 1f | |
| CNNM2/rs12221064 | 2b | |
| NT5C2/rs12412038 | 1f | |
| CNNM2/rs12411886 | 1f | |
| NT5C2/rs12413046 | 1f | |
| CNNM2/rs12413409a | 1f | |
| NT5C2/rs9633712 | 1e | |
| CNNM2/rs17115213 | 1f | |
| NT5C2/rs11191548 | 1f | |
| CNNM2/rs2297787 | 2a | |
| CNNM2/rs3781285 | 1f | |
| CNNM2/rs943037 | 1f | |
| CNNM2/rs12219901 | 2b | |
| APOE-APOC1/TOMM40/rs2075650 | APOE-APOC1/rs2075650a | 1f |
| UBE2Z/rs46522 | GIP/rs2291725 | 1f |
| GIP/rs4794004 | 1d | |
| SNF8/rs1994970 | 1f | |
| SNF8/rs4793992 | 1f | |
| UBE2Z/rs12601672 | 2b | |
| UBE2Z/rs15563 | 1f | |
| UBE2Z/rs3744608 | 2a | |
| UBE2Z/rs3848460 | 1f | |
| UBE2Z/rs46522a | 1f | |
| UBE2Z/rs11079844 | 1f | |
| ZNF259-APOA5-APOA1/rs964184 | ZNF259-APOA5-APOA1/rs964184 | 1f |
| SMG6/rs2281727 | SMG6/rs2281727a | 2b |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| COL4A1-COL4A2/rs4773144 | COL4A1-COL4A2/rs4773144a | 2b |
| ABO/rs579459 | ABO/rs649129 | 2b |
| ADMTS7/rs7173743 | LOC105370915/rs5029904 | 2b |
| PHACTR1/rs4773143 | 2b | |
| CXCL12/rs501120 | CXCL12/rs518594 | 2b |
| CXCL12/rs1746052 | 2b | |
| FURIN-FES/rs17514846 | FES/rs1894401 | 1b |
| HHIPL1/rs2895811 | HHIPL1/rs28391527 | 2b |
| HHIPL1/rs4624107 | 2b | |
| HHIPL1/rs7145262 | 2b | |
| IL6R/rs4845625 | IL6R/rs7549250 | 2b |
| IL6R/rs7549338 | 2b | |
| IL6R/rs7553796 | 2b | |
| KCNE2/rs9982601 | KCNE2/rs28591415 | 2b |
| KIAA1462/rs2505083 | KIAA1462/rs3739998 | 2b |
| LPL/rs264 | LPL/rs271 | 1f |
| LPL/rs3779788 | 2b | |
| MIA3/rs17465637 | MIA3/rs17163301 | 2b |
| PLG/rs4252120 | PLG/rs4252126 | 1f |
| PLG/rs4252135 | 1f | |
| PPAP2B/rs17114036 | LOC101929929/rs72664304 | 2a |
| PLPP3/rs4634932 | 1f | |
| SLC22A4-SLC22A5/rs273909 | SLC22A5/rs17689550 | 1f |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| SORT1/rs602633 | CELSR2/rs12740374 | 2b |
| CELSR2/rs629301 | 1f | |
| CELSR2/rs646776 | 1f | |
| TRIB1/rs2954029 | LOC105375745/rs2980853 | 2b |
| LOC105375745/rs2001844 | 2b | |
| LOC105375745/rs6982636 | 2b | |
| TRIB1/rs2980856 | 2b | |
| VAMP5-VAMP8-GGCX/rs1561198 | GGCX/rs6738645 | 1f |
| GGCX/rs10187424 | 1f | |
| VAMP8/rs1009 | 1b | |
| GGCX/rs6547621 | 1f | |
| VAMP8/rs1348818 | 1f | |
| GGCX/rs2886722 | 1f | |
| VAMP8/rs3770098 | 1f | |
| VAMP8/rs6757263 | 1f | |
| WDR12/rs6725887 | ICA1L/rs72934715 | 2b |
| NBEAL1/rs2351524 | 1f | |
| WDR12/rs72936852 | 2b | |
| NBEAL1/rs4675310 | 1f | |
| NBEAL1/rs72934512 | 2b | |
| REST-NOA1/rs17087335 | REST/rs2227901 | 1f |
| REST-NOA1/rs7687767 | 1d | |
| SWAP70/rs10840293 | SWAP70/rs93138 | 1f |
| SWAP70/rs360136 | 1f | |
| SMAD3/rs56062135 | SMAD3/rs17293632 | 2a |
| SMAD3/rs1866316 | 2b | |
| MTERF1/rs8032739 | 2b | |
| CDKN2BAS1/rs1333049 | CDKN2BAS1/rs4977574 | 2c |
GWAS significant SNPs with functional evidence (RegulomeDB score < 3) are bolded.
Functional SNPs (RegluomeDB Score < 3) in LD (r2 ≥ 0.80) with published GWAS SNPs
| GWAS SNPs . | Functional proxy SNPs . | Regulome DB score . |
|---|---|---|
| LIPA/rs2246833 | LIPA/rs1332327 | 2b |
| LIPA/rs1332328 | 2b | |
| LIPA/rs1412444 | 1d | |
| LIPA/rs2246833a | 1b | |
| LIPA/rs2250644 | 2b | |
| ZC3HC1/rs11556924 | ZC3HC1/rs11556924a | 1f |
| CYP17A1-CNNM2-NT5C2/rs12413409 | AS3MT/rs11191454 | 1f |
| BORCS7-ASMT/rs4409766 | 1f | |
| CNNM2/rs10883808 | 1f | |
| MAT2A/rs1446668 | 2a | |
| NT5C2/rs10883832 | 1f | |
| CNNM2/rs11191479 | 1f | |
| NT5C2/rs11191557 | 1f | |
| CNNM2/rs11191499 | 1f | |
| NT5C2/rs11191558 | 1f | |
| CNNM2/rs11191514 | 1f | |
| NT5C2/rs11191580 | 1f | |
| CNNM2/rs11191515 | 1f | |
| NT5C2/rs11191582 | 1f | |
| CNNM2/rs12221064 | 2b | |
| NT5C2/rs12412038 | 1f | |
| CNNM2/rs12411886 | 1f | |
| NT5C2/rs12413046 | 1f | |
| CNNM2/rs12413409a | 1f | |
| NT5C2/rs9633712 | 1e | |
| CNNM2/rs17115213 | 1f | |
| NT5C2/rs11191548 | 1f | |
| CNNM2/rs2297787 | 2a | |
| CNNM2/rs3781285 | 1f | |
| CNNM2/rs943037 | 1f | |
| CNNM2/rs12219901 | 2b | |
| APOE-APOC1/TOMM40/rs2075650 | APOE-APOC1/rs2075650a | 1f |
| UBE2Z/rs46522 | GIP/rs2291725 | 1f |
| GIP/rs4794004 | 1d | |
| SNF8/rs1994970 | 1f | |
| SNF8/rs4793992 | 1f | |
| UBE2Z/rs12601672 | 2b | |
| UBE2Z/rs15563 | 1f | |
| UBE2Z/rs3744608 | 2a | |
| UBE2Z/rs3848460 | 1f | |
| UBE2Z/rs46522a | 1f | |
| UBE2Z/rs11079844 | 1f | |
| ZNF259-APOA5-APOA1/rs964184 | ZNF259-APOA5-APOA1/rs964184 | 1f |
| SMG6/rs2281727 | SMG6/rs2281727a | 2b |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| COL4A1-COL4A2/rs4773144 | COL4A1-COL4A2/rs4773144a | 2b |
| ABO/rs579459 | ABO/rs649129 | 2b |
| ADMTS7/rs7173743 | LOC105370915/rs5029904 | 2b |
| PHACTR1/rs4773143 | 2b | |
| CXCL12/rs501120 | CXCL12/rs518594 | 2b |
| CXCL12/rs1746052 | 2b | |
| FURIN-FES/rs17514846 | FES/rs1894401 | 1b |
| HHIPL1/rs2895811 | HHIPL1/rs28391527 | 2b |
| HHIPL1/rs4624107 | 2b | |
| HHIPL1/rs7145262 | 2b | |
| IL6R/rs4845625 | IL6R/rs7549250 | 2b |
| IL6R/rs7549338 | 2b | |
| IL6R/rs7553796 | 2b | |
| KCNE2/rs9982601 | KCNE2/rs28591415 | 2b |
| KIAA1462/rs2505083 | KIAA1462/rs3739998 | 2b |
| LPL/rs264 | LPL/rs271 | 1f |
| LPL/rs3779788 | 2b | |
| MIA3/rs17465637 | MIA3/rs17163301 | 2b |
| PLG/rs4252120 | PLG/rs4252126 | 1f |
| PLG/rs4252135 | 1f | |
| PPAP2B/rs17114036 | LOC101929929/rs72664304 | 2a |
| PLPP3/rs4634932 | 1f | |
| SLC22A4-SLC22A5/rs273909 | SLC22A5/rs17689550 | 1f |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| SORT1/rs602633 | CELSR2/rs12740374 | 2b |
| CELSR2/rs629301 | 1f | |
| CELSR2/rs646776 | 1f | |
| TRIB1/rs2954029 | LOC105375745/rs2980853 | 2b |
| LOC105375745/rs2001844 | 2b | |
| LOC105375745/rs6982636 | 2b | |
| TRIB1/rs2980856 | 2b | |
| VAMP5-VAMP8-GGCX/rs1561198 | GGCX/rs6738645 | 1f |
| GGCX/rs10187424 | 1f | |
| VAMP8/rs1009 | 1b | |
| GGCX/rs6547621 | 1f | |
| VAMP8/rs1348818 | 1f | |
| GGCX/rs2886722 | 1f | |
| VAMP8/rs3770098 | 1f | |
| VAMP8/rs6757263 | 1f | |
| WDR12/rs6725887 | ICA1L/rs72934715 | 2b |
| NBEAL1/rs2351524 | 1f | |
| WDR12/rs72936852 | 2b | |
| NBEAL1/rs4675310 | 1f | |
| NBEAL1/rs72934512 | 2b | |
| REST-NOA1/rs17087335 | REST/rs2227901 | 1f |
| REST-NOA1/rs7687767 | 1d | |
| SWAP70/rs10840293 | SWAP70/rs93138 | 1f |
| SWAP70/rs360136 | 1f | |
| SMAD3/rs56062135 | SMAD3/rs17293632 | 2a |
| SMAD3/rs1866316 | 2b | |
| MTERF1/rs8032739 | 2b | |
| CDKN2BAS1/rs1333049 | CDKN2BAS1/rs4977574 | 2c |
| GWAS SNPs . | Functional proxy SNPs . | Regulome DB score . |
|---|---|---|
| LIPA/rs2246833 | LIPA/rs1332327 | 2b |
| LIPA/rs1332328 | 2b | |
| LIPA/rs1412444 | 1d | |
| LIPA/rs2246833a | 1b | |
| LIPA/rs2250644 | 2b | |
| ZC3HC1/rs11556924 | ZC3HC1/rs11556924a | 1f |
| CYP17A1-CNNM2-NT5C2/rs12413409 | AS3MT/rs11191454 | 1f |
| BORCS7-ASMT/rs4409766 | 1f | |
| CNNM2/rs10883808 | 1f | |
| MAT2A/rs1446668 | 2a | |
| NT5C2/rs10883832 | 1f | |
| CNNM2/rs11191479 | 1f | |
| NT5C2/rs11191557 | 1f | |
| CNNM2/rs11191499 | 1f | |
| NT5C2/rs11191558 | 1f | |
| CNNM2/rs11191514 | 1f | |
| NT5C2/rs11191580 | 1f | |
| CNNM2/rs11191515 | 1f | |
| NT5C2/rs11191582 | 1f | |
| CNNM2/rs12221064 | 2b | |
| NT5C2/rs12412038 | 1f | |
| CNNM2/rs12411886 | 1f | |
| NT5C2/rs12413046 | 1f | |
| CNNM2/rs12413409a | 1f | |
| NT5C2/rs9633712 | 1e | |
| CNNM2/rs17115213 | 1f | |
| NT5C2/rs11191548 | 1f | |
| CNNM2/rs2297787 | 2a | |
| CNNM2/rs3781285 | 1f | |
| CNNM2/rs943037 | 1f | |
| CNNM2/rs12219901 | 2b | |
| APOE-APOC1/TOMM40/rs2075650 | APOE-APOC1/rs2075650a | 1f |
| UBE2Z/rs46522 | GIP/rs2291725 | 1f |
| GIP/rs4794004 | 1d | |
| SNF8/rs1994970 | 1f | |
| SNF8/rs4793992 | 1f | |
| UBE2Z/rs12601672 | 2b | |
| UBE2Z/rs15563 | 1f | |
| UBE2Z/rs3744608 | 2a | |
| UBE2Z/rs3848460 | 1f | |
| UBE2Z/rs46522a | 1f | |
| UBE2Z/rs11079844 | 1f | |
| ZNF259-APOA5-APOA1/rs964184 | ZNF259-APOA5-APOA1/rs964184 | 1f |
| SMG6/rs2281727 | SMG6/rs2281727a | 2b |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| COL4A1-COL4A2/rs4773144 | COL4A1-COL4A2/rs4773144a | 2b |
| ABO/rs579459 | ABO/rs649129 | 2b |
| ADMTS7/rs7173743 | LOC105370915/rs5029904 | 2b |
| PHACTR1/rs4773143 | 2b | |
| CXCL12/rs501120 | CXCL12/rs518594 | 2b |
| CXCL12/rs1746052 | 2b | |
| FURIN-FES/rs17514846 | FES/rs1894401 | 1b |
| HHIPL1/rs2895811 | HHIPL1/rs28391527 | 2b |
| HHIPL1/rs4624107 | 2b | |
| HHIPL1/rs7145262 | 2b | |
| IL6R/rs4845625 | IL6R/rs7549250 | 2b |
| IL6R/rs7549338 | 2b | |
| IL6R/rs7553796 | 2b | |
| KCNE2/rs9982601 | KCNE2/rs28591415 | 2b |
| KIAA1462/rs2505083 | KIAA1462/rs3739998 | 2b |
| LPL/rs264 | LPL/rs271 | 1f |
| LPL/rs3779788 | 2b | |
| MIA3/rs17465637 | MIA3/rs17163301 | 2b |
| PLG/rs4252120 | PLG/rs4252126 | 1f |
| PLG/rs4252135 | 1f | |
| PPAP2B/rs17114036 | LOC101929929/rs72664304 | 2a |
| PLPP3/rs4634932 | 1f | |
| SLC22A4-SLC22A5/rs273909 | SLC22A5/rs17689550 | 1f |
| SMG6/rs7217687 | 2b | |
| SMG6/rs9908888 | 2b | |
| SORT1/rs602633 | CELSR2/rs12740374 | 2b |
| CELSR2/rs629301 | 1f | |
| CELSR2/rs646776 | 1f | |
| TRIB1/rs2954029 | LOC105375745/rs2980853 | 2b |
| LOC105375745/rs2001844 | 2b | |
| LOC105375745/rs6982636 | 2b | |
| TRIB1/rs2980856 | 2b | |
| VAMP5-VAMP8-GGCX/rs1561198 | GGCX/rs6738645 | 1f |
| GGCX/rs10187424 | 1f | |
| VAMP8/rs1009 | 1b | |
| GGCX/rs6547621 | 1f | |
| VAMP8/rs1348818 | 1f | |
| GGCX/rs2886722 | 1f | |
| VAMP8/rs3770098 | 1f | |
| VAMP8/rs6757263 | 1f | |
| WDR12/rs6725887 | ICA1L/rs72934715 | 2b |
| NBEAL1/rs2351524 | 1f | |
| WDR12/rs72936852 | 2b | |
| NBEAL1/rs4675310 | 1f | |
| NBEAL1/rs72934512 | 2b | |
| REST-NOA1/rs17087335 | REST/rs2227901 | 1f |
| REST-NOA1/rs7687767 | 1d | |
| SWAP70/rs10840293 | SWAP70/rs93138 | 1f |
| SWAP70/rs360136 | 1f | |
| SMAD3/rs56062135 | SMAD3/rs17293632 | 2a |
| SMAD3/rs1866316 | 2b | |
| MTERF1/rs8032739 | 2b | |
| CDKN2BAS1/rs1333049 | CDKN2BAS1/rs4977574 | 2c |
GWAS significant SNPs with functional evidence (RegulomeDB score < 3) are bolded.
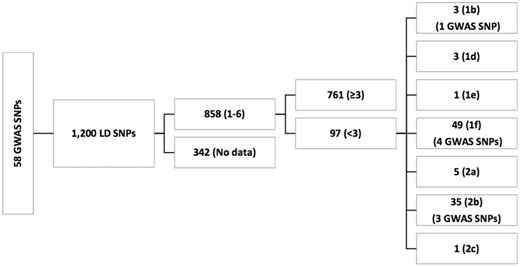
58 GWAS ANPs in LD with 1200 SNPs. We used SNAP webportal to determine LD SNPs. These 1200 SNPs were further evaluated by RegulomeDB to identify their functional role. RegulomeDB did not provide data for 342 SNPs. A total of 858 SNPs returned the scores of 1–6 by RegulomeDB. Of those 858 SNPs, 97 returned the scores of < 3. Among 97 functional SNPs, only 8 were GWAS SNPs. Lower the RegulomeDB score, more evidence of functionality.
Overall, we had 97 functional SNPs (RegulomeDB < 3). Eight of these were GWAS SNPs, and the remaining 89 were in LD (r2 ≥ 0.80) with the GWAS SNPs.
Three variants, FES/rs1894401, LIPA/rs2246833 and VAMP8/rs1009, were strongly predicted to be functional with score of 1b. FES/rs1894401 is an intronic SNP that is an eQTL for FES in thyroid and transformed lymphoblasts, is present in the binding motif of Pax5, and affects the binding of eleven transcription factors. LIPA/rs2246833 (RegulomeDB score = 1b), located in Intron 6 of LIPA, in the DNA motif of EWSRCFLI1, is a GWAS reported SNP along with 4 other functional SNPs (of 12 tested) in this region and it and it affects the binding of CTCF. It is an eQTL in the whole blood. VAMP5-VAMP8-GGCX/rs1009 is in exon 3 of VAMP8 and affects the binding of CTCF and HSF1. rs1009 of VAMP8 is an eQTL in lymhoblasts, skeletal muscles, adipose tissue and thyroid. Of 42 SNPs analyzed in this locus, we found 8 other SNPs with RegulomeDB score < 3 (Table 3).
There were 33 functional SNPs within 15 GWAS identified CAD loci: ABO (1of 10 assessed), ADAMTS7 (1 of 15 assessed), CXCL12 (2 of 36 assessed), HHIPL1 (3 of 17 assessed), KCNE2(2 of 18 assessed), KIAA1462 (1 of 9 assessed), MIA3 (1 of 27 assessed), PPAP2B (2 of 22 assessed), SORT1 (3 of 9 assessed), WDR12 (5 of 214 assessed), IL6R (3 of 14 assessed), LPL (2 of 6 assessed), PLG (2 of 41 assessed), SLC22A4-SLC22A5 (1 of 2 assessed) and TRIB1 (4 of 16 assessed).
Of 97 SNPs with RegulomeDB score < 3, 25 were in the CYP17A1-CNNM2-NT5C2 region, and one of them was a GWAS reported SNP (rs12413409). The regional LD plot of this SNP is given in Supplementary Figure S1. rs9633712 (RegulomeDB score = 1e) is located in Intron 3 of NT5C2 and is an eQTL for USMG5 in monocytes. This SNP was also found in the motifs of the following transcription factors: PU1, ELF1, Sfpil, PU.1 and c-Ets-1. It appears to affect the binding of SPI1. Twenty SNPs returned a score of 1f (likely to affect the binding), and 18 of them were in intronic regions. NT5C2/rs11191558 lies in HOXC series of DNA motifs, and CNNM2/rs3781285 lies between NF-kappaB and P50:50. NT5C2/rs2297787 returned a score of 2a, affecting the binding motifs of FOXI1, HNF3-alpha and FOXP1 and the binding of FOXA1. SNP rs12412038 is located in Intron 10 of NT5C2 and is in the binding motif of Irx. The remaining two SNPs, rs12219901 and rs12221064, lie in the CNNM2-NT5C2 intergenic region and upstream of CNNM2, respectively. They are located in DNA motifs of SRF and MAZR and affect the binding of POLRA2 and CTCF/ETS. Interestingly, rs943037 resides in exon 7 of CNNM2. Nineteen of the 25 SNPs in the region of CYP17A1-CNNM2-NT5C2 are eQTLs for USMG5 (Table 4).
Putative functional SNPs and corresponding motifs, eQTL and related transcription factors (Regulome DB score < 3)
| Coordinate 0-based . | SNP ID . | RegulomeDB score . | Gene/Locus . | Position . | eQTL . | Motif . | Protein-binding . | |
|---|---|---|---|---|---|---|---|---|
| chr15:91429041 | rs1894401 | 1b | FES | Intron 2 | FES | Pax5 | SPI1 | |
| USF1 | ||||||||
| POLR2A | ||||||||
| GABPA | ||||||||
| BHLHE40 | ||||||||
| CEBPB | ||||||||
| CTCF | ||||||||
| MAX | ||||||||
| RFX5 | ||||||||
| RUNX3 | ||||||||
| STAT5A | ||||||||
| chr10:91005853 | rs2246833 | 1b | LIPA | Intron 6 | LIPA | EWSR-FLI1 | CTCF | |
| znf143 | ||||||||
| chr2:85808736 | rs1009 | 1b | VAMP8 | Exon 3 | VAMP8 | CTCF | ||
| LOC388969 | HSF1 | |||||||
| chr17:47038470 | rs4794004 | 1d | GIP | Intron 4 | ATP5G1 | Gata5 | NR3C1 | |
| UBE2Z | IN3AK20 | |||||||
| CREB1 | ||||||||
| TAF1 | ||||||||
| TCF12 | ||||||||
| CTCF | ||||||||
| POLR2A | ||||||||
| USF1 | ||||||||
| FOXA1 | ||||||||
| FOXA2 | ||||||||
| RBBP5 | ||||||||
| chr10:91002926 | rs1412444 | 1d | LIPA | Intron3 | LIPA | SAP1a | ATF2 | |
| ELK1 | FOXM1 | |||||||
| ELK3 | SP1 | |||||||
| ELK4 | SPI1 | |||||||
| MECP2 | MTA3 | |||||||
| ERF | RUNX3 | |||||||
| ERG | ||||||||
| ETS1 | ||||||||
| ETV1 | ||||||||
| ETV2 | ||||||||
| ETV3 | ||||||||
| Gabpa | ||||||||
| chr10:104873760 | rs9633712 | 1e | NT5C2 | Intron 3 | UMG5 | PU1 | SP11 | |
| ELF-1 | ||||||||
| Sfpil | ||||||||
| PU.1 | ||||||||
| c-Ets-1 | ||||||||
| chr11:116648916 | rs964184 | 1f | ZPR1 | Downstream ZRP1 | TAGLN | FOXJ2 | ||
| chr1:109818529 | rs646776 | 1f | CELSR2 | Upstream CELSR2 | PSMA5 | CTCF | ||
| HEY1 | ||||||||
| REST | ||||||||
| POLR2A | ||||||||
| ZBTB7A | ||||||||
| TAF7 | ||||||||
| chr10:104616662 | rs4409766 | 1f | BORCS7-ASMT | Intron 1 | C10orf77 | Tcf3 | BACH1 | |
| USMG5 | MAFF | |||||||
| MAFK | ||||||||
| chr17:47008206 | rs4793992 | 1f | SNF8 | Intron 7 | ATP5G1 | POLR2A | ||
| UBE2Z | TEAD4 | |||||||
| chr6:161152293 | rs4252126 | 1f | PLG | Intron 11 | PLG | CTCF | ||
| RUNX3 | ||||||||
| TEAD4 | ||||||||
| RAD21 | ||||||||
| chr6:161154231 | rs4252135 | 1f | PLG | Intron 12 | PLG | CTCF | ||
| FOXA1 | ||||||||
| NFKB1 | ||||||||
| RAD21 | ||||||||
| ZNF263 | ||||||||
| SMC3 | ||||||||
| ZNF143 | ||||||||
| FOXA2 | ||||||||
| chr10:104846177 | rs11191548 | 1f | NT5C2 gene region | Downstream NT5C2 | USMG5 | TEAD1 | ||
| TEAD3 | ||||||||
| chr10:104864613 | rs11191557 | 1f | NT5C2 | Intron 5 | USMG5 | |||
| chr10:104864677 | rs11191558 | 1f | NT5C2 | Intron 5 | USMG5 | HOXC13 | ||
| Hoxa13 | ||||||||
| Hoxc13 | ||||||||
| Hoxd12 | ||||||||
| HOXA13 | ||||||||
| HOXD9 | ||||||||
| HOXC11 | ||||||||
| chr10:104871203 | rs12413046 | 1f | NT5C2 | Intron 3 | USMG5 | NR3C1 | ||
| TRIM28 | ||||||||
| CTCF | ||||||||
| ATF2 | ||||||||
| IKZF1 | ||||||||
| TCF7L2 | ||||||||
| ZNF263 | ||||||||
| chr10:104871278 | rs10883832 | 1f | NT5C2 | Intron 3 | USMG5 | TRIM28 | ||
| TCF7L2 | ||||||||
| chr10:104913652 | rs11191582 | 1f | NT5C2 | Intron 2 | USMG5 | EP300 | ||
| NFIC | ||||||||
| TCF12 | ||||||||
| TEAD4 | ||||||||
| STAT1 | ||||||||
| ARID3A | ||||||||
| EP300 | ||||||||
| JUN | ||||||||
| RCOR1 | ||||||||
| chr10:104906210 | rs11191580 | 1f | NT5C2 | Intron 2 | USMG5 | TRIM28 | ||
| SETDB1 | ||||||||
| GATA1 | ||||||||
| GTF2F1 | ||||||||
| CEBPB | ||||||||
| FOS | ||||||||
| JUND | ||||||||
| ZNF263 | ||||||||
| chr10:104856161 | rs12412038 | 1f | NT5C2 | Intron 10 | USMG5 | Irx-3 | ||
| Irx-2 | ||||||||
| Irx-4 | ||||||||
| Irx-6 | ||||||||
| chr2:85807081 | rs1348818 | 1f | VAMP8 | Intron 2 | GGCX | HMGIY | EBF1, | |
| Mtf1 | ||||||||
| Srf | ||||||||
| Zfp105 | ||||||||
| HMGIY | ||||||||
| chr2:85805366 | rs3770098 | 1f | VAMP8 | Intron1 | VAMP8 | POLR2A | ||
| LOC388969 | BHLHE40 | |||||||
| E2F6 | ||||||||
| KDM5B | ||||||||
| MAX | ||||||||
| MXI1 | ||||||||
| MYC | ||||||||
| NFIC | ||||||||
| WRNIP1 | ||||||||
| chr2:85803541 | rs6757263 | 1f | VAMP8 | Upstream VAMP8 | GGCX | SP1 | ||
| VAMP8 | EP300 | |||||||
| LOC388969 | NFIC | |||||||
| chr17:46988596 | rs46522 | 1f | UBE2Z | Intron 2 | ATP5G1 | NFKB | ||
| UBE2Z | NFYB | |||||||
| RUNX3 | ||||||||
| chr19:45395618 | rs2075650 | 1f | TOMM40 | Intron 2 | TOMM40 | RREB1 | ||
| chr1:56996190 | rs4634932 | 1f | PLPP3 | Intron 2 | PPAP2B | POLR2A | ||
| chr2:203880833 | rs4675310 | 1f | NBEAL1 | Intron 1 | ALS2CR13 | |||
| chr10:104681142 | rs17115213 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104721125 | rs10883808 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104723619 | rs11191479 | 1f | CNNM2 | Intron 1 | USMG5 | GATA1 | ||
| TAL1 | ||||||||
| CEBPB | ||||||||
| chr10:104773363 | rs11191514 | 1f | CNNM2 | Intron 1 | USMG5 | PAX5 | ||
| chr10:104776526 | rs11191515 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104825664 | rs3781285 | 1f | CNNM2 | Intron 4 | USMG5 | NF-kappaB | IKZF1 | |
| P50:50 | ||||||||
| chr10:104835918 | rs943037 | 1f | CNNM2 | Exon 7 | USMG5 | TBX20 | ||
| Foxj1 | ||||||||
| chr8:19813701 | rs271 | 1f | LPL | Intron 6 | LPL | |||
| chr17:47039131 | rs2291725 | 1f | GIP | Exon 4 | GIP | GATA2 | ||
| TCF4 | ||||||||
| FOSL2 | ||||||||
| EGR1 | ||||||||
| ELF1 | ||||||||
| FOS | ||||||||
| NR3C1 | ||||||||
| EP300 | ||||||||
| RXRA | ||||||||
| CHD2 | ||||||||
| JUND | ||||||||
| POLR2A | ||||||||
| RAD21 | ||||||||
| FOSL1 | ||||||||
| REST | ||||||||
| chr2:203880991 | rs2351524 | 1f | NBEAL1 | 5' UTR | ALS2CR13 | |||
| chr1:109818305 | rs629301 | 1f | CELSR2 | 3' UTR | PSRC1 | CTCF | ||
| POLR2A | ||||||||
| chr2:85774675 | rs6547621 | 1f | 3' UTR | GGCX | ELK4 | |||
| POLR2A | ||||||||
| chr10:104660003 | rs11191454 | 1f | AS3MT | Intron10 | USMG5 | |||
| chr10:104685298 | rs12411886 | 1f | CNNM2 | Intron1 | USMG5 | Zec | ||
| chr10:104719095 | rs12413409 | 1f | CNNM2 | Intron1 | USMG5 | POLR3A | ||
| chr10:104764270 | rs11191499 | 1f | CNNM2 | Intron1 | USMG5 | |||
| chr17:47014126 | rs1994970 | 1f | SNF8 | Intron4 | ATP5G1 | TFII-I | ||
| UBE2Z | ||||||||
| chr2:85742296 | rs2886722 | 1f | Pseudogene | LOC388969 | TCF7L2 | |||
| chr2:85783127 | rs6738645 | 1f | GGCX | Intron5 | Evi-1 | POLR2A | ||
| chr2:85794296 | rs10187424 | 1f | Pseudogene | GGCX | ||||
| LOC388969 | ||||||||
| chr5:131723064 | rs17689550 | 1f | RAPGEF6 | |||||
| chr7:129663495 | rs11556924 | 1f | ZC3HC1 | Exon8 | KIAA0265 | |||
| chr17:4702833 | rs11079844 | 1f | Pseudogene | ATP5G1 | ||||
| chr17: 47005192 | rs15563 | 1f | UBE2Z | Exon7 | ATP5G1 | PRDM1 | ||
| chr17:47047113 | rs3848460 | 1f | UBE2Z | ATP5G1 | CEBPB | |||
| chr10:104680136 | rs2297787 | 2a | CNNM2 | Intron 1 | Freac-7 | FOXA1 | ||
| HFH3(FOXI1) | SIN3A | |||||||
| HNF3alpha | ZNF263 | |||||||
| FOXP1 | HNF4G | |||||||
| Elf3 | FOXA1 | |||||||
| Foxl1 | ||||||||
| Srf | ||||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| Zfp105 | ||||||||
| HFH(FOXl1) | ||||||||
| chr1:56948289 | rs72664304 | 2a | C8B | Intron 6 | FOXA1 | FOXA1 | ||
| Foxa2 | FOXA2 | |||||||
| TCF4 | ||||||||
| SP1 | ||||||||
| HNF4G | ||||||||
| HNF4A | ||||||||
| HDAC2 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| chr17:46993232 | rs3744608 | 2a | UBE2Z | Intron 3 | Zfp740 | SPI1 | ||
| MZF1 | POLR2A | |||||||
| MAZR | IKZF1 | |||||||
| SP1 | MAX | |||||||
| SP1:SP3 | TFAP2A | |||||||
| WT1ZNF21 | TFAP2C | |||||||
| Zfp281 | SP1 | |||||||
| ZFp740 | CEBPB | |||||||
| WT1 | NR3C1 | |||||||
| ZNF219 | BATF | |||||||
| ZNF740 | BCL11A | |||||||
| SP4 | MEF2A | |||||||
| NFKB1 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| STAT3 | ||||||||
| IRF4 | ||||||||
| EBF1 | ||||||||
| FOSL2 | ||||||||
| BATF | ||||||||
| NR3C1 | ||||||||
| RUNX3 | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr2:85764959 | rs1446668 | 2a | MAT2A | nc transcript | CTCF | CTCF | ||
| Upstream MAT2A | POLR2A | |||||||
| TAF1 | ||||||||
| RFX5 | ||||||||
| RAD21 | ||||||||
| HEY1 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| ZNF263 | ||||||||
| NR3C1 | ||||||||
| CTCF | ||||||||
| MYC | ||||||||
| AR | ||||||||
| MYBL2 | ||||||||
| TEAD4 | ||||||||
| MAZ | ||||||||
| CHD2 | ||||||||
| SMC3 | ||||||||
| TBP | ||||||||
| ZNF143 | ||||||||
| CDX2 | ||||||||
| E2F6 | ||||||||
| MAX | ||||||||
| NR3C1 | ||||||||
| SIN3A | ||||||||
| YY1 | ||||||||
| REST | ||||||||
| HMGN3 | ||||||||
| chr17:47006492 | rs12601672 | 2b | UBE2Z | Downstream UBE2Z | Zfx | POLR2A | ||
| EGR1 | ||||||||
| SPI1 | ||||||||
| ELF1 | ||||||||
| chr10:30316071 | rs3739998 | 2b | KIAA1462 | Exon 2 | RELA | CTCF | ||
| MYC | ||||||||
| PAX5 | ||||||||
| ZNF143 | ||||||||
| chr8:126476378 | rs2980856 | 2b | TRIB1 gene | Intergenic region | pax-8 | JUND | ||
| region | Downstream TRIB1 | Sox17 | POLR2A | |||||
| TFAP2C | ||||||||
| MXI1 | ||||||||
| CEBPB | ||||||||
| chr9:136154303 | rs649129 | 2b | ABO gene region | Intergenic region | IRF | NFYA | ||
| Upstream ABO | POLR2A | |||||||
| FOS | ||||||||
| IRF1 | ||||||||
| NFYB | ||||||||
| PML | ||||||||
| chr10:104840966 | rs12219901 | 2b | CNNM2 gene | Intergenic region | SRF | POLR2A | ||
| region | Downstream CXCL12 | |||||||
| chr10:44778545 | rs1746052 | 2b | CXCL12 gene | Intergenic region | GATA1 | TAL1 | ||
| region | Downstream CXCL12 | |||||||
| chr21:35593826 | rs28451064 | 2b | LINC00310 gene | Intergenic region | PPAR | SP1 | ||
| region | Downstream | FOXA2 | ||||||
| LINC00310 | ||||||||
| chr17:2098271 | rs7217687 | 2b | SMG6 | Intron 13 | - | NF-1 | SIN3A | |
| TCF12 | ||||||||
| MAX | ||||||||
| YY1 | ||||||||
| ZNF263 | ||||||||
| EP300 | ||||||||
| TEAD4 | ||||||||
| chr13:110960711 | rs4773144 | 2b | COL4A2 | Intron 3 | STAT3:STAT3 | POLR2A | ||
| EZH2 | ||||||||
| chr14:100116251 | rs28391527 | 2b | HHIPL1 | Intron 3 | MyoD | BHLHE40 | ||
| SCRT1 | USF1 | |||||||
| FIGLA | FOXA1 | |||||||
| MAX | ||||||||
| chr1:154404335 | rs7549250 | 2b | IL6R | Intron 3 | TBX15 | MXI1 | ||
| FOS | ||||||||
| JUNB | ||||||||
| MAX | ||||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| FOSL1 | ||||||||
| MAFK | ||||||||
| RCOR1 | ||||||||
| MYC | ||||||||
| USF2 | ||||||||
| TEAD4 | ||||||||
| RCOR1 | ||||||||
| YY1 | ||||||||
| chr1:154404379 | rs7549338 | 2b | IL6R | Intron 3 | GR | FOS | ||
| AR | JUNB | |||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr1:154404405 | rs7553796 | 2b | IL6R | Intron 3 | NF-kappaB, | FOS | ||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr14:100127439 | rs4624107 | 2b | HHIPL1 | Intron 7 | Pax5 | JUND | ||
| chr10:91011457 | rs1332328 | 2b | LIPA | Intron 9 | UF1H3BETA | CREBBP | ||
| ZNF263 | ||||||||
| CDX2 | ||||||||
| ELF1 | ||||||||
| ZEB1 | ||||||||
| TBP | ||||||||
| TFAPC2 | ||||||||
| TBP | ||||||||
| POLR2A | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| HEY1 | ||||||||
| chr17:2117944 | rs2281727 | 2b | SMG6 | Intron 13 | SRY | CREBBP | ||
| Srf | EP300 | |||||||
| Zfp105 | STAT3 | |||||||
| TRIM28 | ||||||||
| MYC | ||||||||
| RBBP5 | ||||||||
| chr8:126479314 | rs6982636 | 2b | LOC105375745 | Intron1 | MAF | SMARCC1 | ||
| RFX3 | ||||||||
| POLR2A | ||||||||
| GATA2 | ||||||||
| CHD2 | ||||||||
| GTF2F1 | ||||||||
| chr14:100125720 | rs7145262 | 2b | HHIPL1 | Intron4 | ESR2 | SMARC4 | ||
| ZBTB7A | ||||||||
| SMARB1 | ||||||||
| POLR2A | ||||||||
| EZH2 | ||||||||
| RAD21 | ||||||||
| BACH1 | ||||||||
| chr10:104677125 | rs12221064 | 2b | CNNM2 | Upstream CNNM2 | MAZR, | CTCF, ETS1 | ||
| ETS1 | ||||||||
| chr8:126478349 | rs2980853 | 2b | LOC105375745 | Upstream | Pit-1, | RFX3 | ||
| LOC105375745 | ||||||||
| chr15:79152421 | rs5029904 | 2b | LOC105370915 | Upstream | NeuroD | USF1 | ||
| LOC105370915 | POLR2A | |||||||
| YY1 | ||||||||
| FOXA1 | ||||||||
| E2F4 | ||||||||
| MAX | ||||||||
| TAF7 | ||||||||
| TAF1 | ||||||||
| MXI1 | ||||||||
| chr8:126478744 | rs2001844 | 2b | LOC105375745 | Upstream | HSF1 | RFX3 | ||
| LOC105375745 | ||||||||
| chr10:91011680 | rs1332327 | 2b | LIPA | 5' UTR | AP-4, | CREBBP | ||
| CDX2 | ||||||||
| ELF1 | ||||||||
| TBP | ||||||||
| SPI1 | ||||||||
| NRF1 | ||||||||
| ELF1 | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| SPI1 | ||||||||
| PAX5 | ||||||||
| SREBF1 | ||||||||
| chr17:2102452 | rs9908888 | 2b | SMG6 | Intron10 | GR | CEBPB | ||
| AR | ||||||||
| chr10:91008878 | rs2250644 | 2b | LIPA | Intron1 | Oct-1 | RUNX3 | ||
| XBP-1 | ||||||||
| MAfb | ||||||||
| Mafk | ||||||||
| MAFB | ||||||||
| MAFK | ||||||||
| NRL | ||||||||
| chr1:109817589 | rs12740374 | 2b | CELSR2 | Exon34 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| DUXA | ||||||||
| chr1:222794090 | rs17163301 | 2b | MIA3 | Intron1 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| HNF1B | ||||||||
| DUXA | ||||||||
| chr21:35644028 | rs28591415 | 2b | pseudogene | PPAR | EP300 | |||
| FOXA1 | ||||||||
| HDAC2 | ||||||||
| NFIC | ||||||||
| SP1 | ||||||||
| chr2:203713279 | rs72934715 | 2b | ICA1L | Intron2 | HMGIY | ATF2 | ||
| NFIC | ||||||||
| EBF1 | ||||||||
| EP300 | ||||||||
| NFKB1 | ||||||||
| PAX5 | ||||||||
| chr2:203775474 | rs72936852 | 2b | WDR12 | Intron1 | AR | MAFF | ||
| chr2:203926270 | rs72934512 | 2b | NBEAL1 | Intron6 | TEAD1 | TEAD4 | ||
| TEAD3 | ||||||||
| chr8:19803092 | rs3779788 | 2b | LPL | Intron1 | TGIF | CEBPB | ||
| chr10:44757106 | rs518594 | 2b | CXCL12 | Downstream intergenic | E2A | FOXM1 | ||
| NRSE | NFIC | |||||||
| NRSF | MAX | |||||||
| EBF1 | ||||||||
| TBL1XR1 | ||||||||
| chr4:57824931 | rs7687767 | 1d | CECR6 | Sox8 | FOXA1 | |||
| chr11:5759712 | rs93138 | 1f | SWAP70 | Intron8 | ||||
| rs360136 | 1f | SWAP70 | Exon13 | |||||
| chr:457798188 | rs2227901 | 1f | REST-NOA1 | Exon6 | SPI1 | |||
| chr:1567442595 | rs17293632 | 2a | SMAD3 | Intron4 | Bach1 | SIN3A | ||
| AP-1 | TCF7L2 | |||||||
| JundM2 | TFAP2A | |||||||
| Pou1f1 | TFAP2C | |||||||
| Pou3f1 | ZNF217 | |||||||
| Sox5 | POLR2A | |||||||
| JDP2 | STAT1 | |||||||
| MXI1 | ||||||||
| TAF1 | ||||||||
| E2F1 | ||||||||
| POLR2A | ||||||||
| HDAC1 | ||||||||
| TCF7L2 | ||||||||
| MYC | ||||||||
| POLR2A | ||||||||
| ESR1 | ||||||||
| EP300 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| CEBPB | ||||||||
| EP300 | ||||||||
| FOS | ||||||||
| FOSL1 | ||||||||
| GATA3 | ||||||||
| JUNB | ||||||||
| JUN | ||||||||
| MYC | ||||||||
| NR2F2 | ||||||||
| NR3C1 | ||||||||
| RAD21 | ||||||||
| RCOR1 | ||||||||
| TCF12 | ||||||||
| JUND | ||||||||
| MAFK | ||||||||
| EGR1 | ||||||||
| MAX | ||||||||
| chr15:67441996 | rs18866316 | 2b | SMAD3 | Exon4 | AIRE | ESR1 | ||
| Elf3 | NR3C1 | |||||||
| Srf | POLR2A | |||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| chr15:67448898 | rs8032739 | 2b | MTERF | Intron4 | SRF | CEBPB | ||
| FOS | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr9:22098573 | rs4977574 | 2c | CDKN2BAS1 | Intron16 | AR | AR | ||
| NR3C1 | ||||||||
| NR3C2 | ||||||||
| Ar | ||||||||
| Elk-1 | ||||||||
| c-Ets-1(p54) | ||||||||
| Coordinate 0-based . | SNP ID . | RegulomeDB score . | Gene/Locus . | Position . | eQTL . | Motif . | Protein-binding . | |
|---|---|---|---|---|---|---|---|---|
| chr15:91429041 | rs1894401 | 1b | FES | Intron 2 | FES | Pax5 | SPI1 | |
| USF1 | ||||||||
| POLR2A | ||||||||
| GABPA | ||||||||
| BHLHE40 | ||||||||
| CEBPB | ||||||||
| CTCF | ||||||||
| MAX | ||||||||
| RFX5 | ||||||||
| RUNX3 | ||||||||
| STAT5A | ||||||||
| chr10:91005853 | rs2246833 | 1b | LIPA | Intron 6 | LIPA | EWSR-FLI1 | CTCF | |
| znf143 | ||||||||
| chr2:85808736 | rs1009 | 1b | VAMP8 | Exon 3 | VAMP8 | CTCF | ||
| LOC388969 | HSF1 | |||||||
| chr17:47038470 | rs4794004 | 1d | GIP | Intron 4 | ATP5G1 | Gata5 | NR3C1 | |
| UBE2Z | IN3AK20 | |||||||
| CREB1 | ||||||||
| TAF1 | ||||||||
| TCF12 | ||||||||
| CTCF | ||||||||
| POLR2A | ||||||||
| USF1 | ||||||||
| FOXA1 | ||||||||
| FOXA2 | ||||||||
| RBBP5 | ||||||||
| chr10:91002926 | rs1412444 | 1d | LIPA | Intron3 | LIPA | SAP1a | ATF2 | |
| ELK1 | FOXM1 | |||||||
| ELK3 | SP1 | |||||||
| ELK4 | SPI1 | |||||||
| MECP2 | MTA3 | |||||||
| ERF | RUNX3 | |||||||
| ERG | ||||||||
| ETS1 | ||||||||
| ETV1 | ||||||||
| ETV2 | ||||||||
| ETV3 | ||||||||
| Gabpa | ||||||||
| chr10:104873760 | rs9633712 | 1e | NT5C2 | Intron 3 | UMG5 | PU1 | SP11 | |
| ELF-1 | ||||||||
| Sfpil | ||||||||
| PU.1 | ||||||||
| c-Ets-1 | ||||||||
| chr11:116648916 | rs964184 | 1f | ZPR1 | Downstream ZRP1 | TAGLN | FOXJ2 | ||
| chr1:109818529 | rs646776 | 1f | CELSR2 | Upstream CELSR2 | PSMA5 | CTCF | ||
| HEY1 | ||||||||
| REST | ||||||||
| POLR2A | ||||||||
| ZBTB7A | ||||||||
| TAF7 | ||||||||
| chr10:104616662 | rs4409766 | 1f | BORCS7-ASMT | Intron 1 | C10orf77 | Tcf3 | BACH1 | |
| USMG5 | MAFF | |||||||
| MAFK | ||||||||
| chr17:47008206 | rs4793992 | 1f | SNF8 | Intron 7 | ATP5G1 | POLR2A | ||
| UBE2Z | TEAD4 | |||||||
| chr6:161152293 | rs4252126 | 1f | PLG | Intron 11 | PLG | CTCF | ||
| RUNX3 | ||||||||
| TEAD4 | ||||||||
| RAD21 | ||||||||
| chr6:161154231 | rs4252135 | 1f | PLG | Intron 12 | PLG | CTCF | ||
| FOXA1 | ||||||||
| NFKB1 | ||||||||
| RAD21 | ||||||||
| ZNF263 | ||||||||
| SMC3 | ||||||||
| ZNF143 | ||||||||
| FOXA2 | ||||||||
| chr10:104846177 | rs11191548 | 1f | NT5C2 gene region | Downstream NT5C2 | USMG5 | TEAD1 | ||
| TEAD3 | ||||||||
| chr10:104864613 | rs11191557 | 1f | NT5C2 | Intron 5 | USMG5 | |||
| chr10:104864677 | rs11191558 | 1f | NT5C2 | Intron 5 | USMG5 | HOXC13 | ||
| Hoxa13 | ||||||||
| Hoxc13 | ||||||||
| Hoxd12 | ||||||||
| HOXA13 | ||||||||
| HOXD9 | ||||||||
| HOXC11 | ||||||||
| chr10:104871203 | rs12413046 | 1f | NT5C2 | Intron 3 | USMG5 | NR3C1 | ||
| TRIM28 | ||||||||
| CTCF | ||||||||
| ATF2 | ||||||||
| IKZF1 | ||||||||
| TCF7L2 | ||||||||
| ZNF263 | ||||||||
| chr10:104871278 | rs10883832 | 1f | NT5C2 | Intron 3 | USMG5 | TRIM28 | ||
| TCF7L2 | ||||||||
| chr10:104913652 | rs11191582 | 1f | NT5C2 | Intron 2 | USMG5 | EP300 | ||
| NFIC | ||||||||
| TCF12 | ||||||||
| TEAD4 | ||||||||
| STAT1 | ||||||||
| ARID3A | ||||||||
| EP300 | ||||||||
| JUN | ||||||||
| RCOR1 | ||||||||
| chr10:104906210 | rs11191580 | 1f | NT5C2 | Intron 2 | USMG5 | TRIM28 | ||
| SETDB1 | ||||||||
| GATA1 | ||||||||
| GTF2F1 | ||||||||
| CEBPB | ||||||||
| FOS | ||||||||
| JUND | ||||||||
| ZNF263 | ||||||||
| chr10:104856161 | rs12412038 | 1f | NT5C2 | Intron 10 | USMG5 | Irx-3 | ||
| Irx-2 | ||||||||
| Irx-4 | ||||||||
| Irx-6 | ||||||||
| chr2:85807081 | rs1348818 | 1f | VAMP8 | Intron 2 | GGCX | HMGIY | EBF1, | |
| Mtf1 | ||||||||
| Srf | ||||||||
| Zfp105 | ||||||||
| HMGIY | ||||||||
| chr2:85805366 | rs3770098 | 1f | VAMP8 | Intron1 | VAMP8 | POLR2A | ||
| LOC388969 | BHLHE40 | |||||||
| E2F6 | ||||||||
| KDM5B | ||||||||
| MAX | ||||||||
| MXI1 | ||||||||
| MYC | ||||||||
| NFIC | ||||||||
| WRNIP1 | ||||||||
| chr2:85803541 | rs6757263 | 1f | VAMP8 | Upstream VAMP8 | GGCX | SP1 | ||
| VAMP8 | EP300 | |||||||
| LOC388969 | NFIC | |||||||
| chr17:46988596 | rs46522 | 1f | UBE2Z | Intron 2 | ATP5G1 | NFKB | ||
| UBE2Z | NFYB | |||||||
| RUNX3 | ||||||||
| chr19:45395618 | rs2075650 | 1f | TOMM40 | Intron 2 | TOMM40 | RREB1 | ||
| chr1:56996190 | rs4634932 | 1f | PLPP3 | Intron 2 | PPAP2B | POLR2A | ||
| chr2:203880833 | rs4675310 | 1f | NBEAL1 | Intron 1 | ALS2CR13 | |||
| chr10:104681142 | rs17115213 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104721125 | rs10883808 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104723619 | rs11191479 | 1f | CNNM2 | Intron 1 | USMG5 | GATA1 | ||
| TAL1 | ||||||||
| CEBPB | ||||||||
| chr10:104773363 | rs11191514 | 1f | CNNM2 | Intron 1 | USMG5 | PAX5 | ||
| chr10:104776526 | rs11191515 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104825664 | rs3781285 | 1f | CNNM2 | Intron 4 | USMG5 | NF-kappaB | IKZF1 | |
| P50:50 | ||||||||
| chr10:104835918 | rs943037 | 1f | CNNM2 | Exon 7 | USMG5 | TBX20 | ||
| Foxj1 | ||||||||
| chr8:19813701 | rs271 | 1f | LPL | Intron 6 | LPL | |||
| chr17:47039131 | rs2291725 | 1f | GIP | Exon 4 | GIP | GATA2 | ||
| TCF4 | ||||||||
| FOSL2 | ||||||||
| EGR1 | ||||||||
| ELF1 | ||||||||
| FOS | ||||||||
| NR3C1 | ||||||||
| EP300 | ||||||||
| RXRA | ||||||||
| CHD2 | ||||||||
| JUND | ||||||||
| POLR2A | ||||||||
| RAD21 | ||||||||
| FOSL1 | ||||||||
| REST | ||||||||
| chr2:203880991 | rs2351524 | 1f | NBEAL1 | 5' UTR | ALS2CR13 | |||
| chr1:109818305 | rs629301 | 1f | CELSR2 | 3' UTR | PSRC1 | CTCF | ||
| POLR2A | ||||||||
| chr2:85774675 | rs6547621 | 1f | 3' UTR | GGCX | ELK4 | |||
| POLR2A | ||||||||
| chr10:104660003 | rs11191454 | 1f | AS3MT | Intron10 | USMG5 | |||
| chr10:104685298 | rs12411886 | 1f | CNNM2 | Intron1 | USMG5 | Zec | ||
| chr10:104719095 | rs12413409 | 1f | CNNM2 | Intron1 | USMG5 | POLR3A | ||
| chr10:104764270 | rs11191499 | 1f | CNNM2 | Intron1 | USMG5 | |||
| chr17:47014126 | rs1994970 | 1f | SNF8 | Intron4 | ATP5G1 | TFII-I | ||
| UBE2Z | ||||||||
| chr2:85742296 | rs2886722 | 1f | Pseudogene | LOC388969 | TCF7L2 | |||
| chr2:85783127 | rs6738645 | 1f | GGCX | Intron5 | Evi-1 | POLR2A | ||
| chr2:85794296 | rs10187424 | 1f | Pseudogene | GGCX | ||||
| LOC388969 | ||||||||
| chr5:131723064 | rs17689550 | 1f | RAPGEF6 | |||||
| chr7:129663495 | rs11556924 | 1f | ZC3HC1 | Exon8 | KIAA0265 | |||
| chr17:4702833 | rs11079844 | 1f | Pseudogene | ATP5G1 | ||||
| chr17: 47005192 | rs15563 | 1f | UBE2Z | Exon7 | ATP5G1 | PRDM1 | ||
| chr17:47047113 | rs3848460 | 1f | UBE2Z | ATP5G1 | CEBPB | |||
| chr10:104680136 | rs2297787 | 2a | CNNM2 | Intron 1 | Freac-7 | FOXA1 | ||
| HFH3(FOXI1) | SIN3A | |||||||
| HNF3alpha | ZNF263 | |||||||
| FOXP1 | HNF4G | |||||||
| Elf3 | FOXA1 | |||||||
| Foxl1 | ||||||||
| Srf | ||||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| Zfp105 | ||||||||
| HFH(FOXl1) | ||||||||
| chr1:56948289 | rs72664304 | 2a | C8B | Intron 6 | FOXA1 | FOXA1 | ||
| Foxa2 | FOXA2 | |||||||
| TCF4 | ||||||||
| SP1 | ||||||||
| HNF4G | ||||||||
| HNF4A | ||||||||
| HDAC2 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| chr17:46993232 | rs3744608 | 2a | UBE2Z | Intron 3 | Zfp740 | SPI1 | ||
| MZF1 | POLR2A | |||||||
| MAZR | IKZF1 | |||||||
| SP1 | MAX | |||||||
| SP1:SP3 | TFAP2A | |||||||
| WT1ZNF21 | TFAP2C | |||||||
| Zfp281 | SP1 | |||||||
| ZFp740 | CEBPB | |||||||
| WT1 | NR3C1 | |||||||
| ZNF219 | BATF | |||||||
| ZNF740 | BCL11A | |||||||
| SP4 | MEF2A | |||||||
| NFKB1 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| STAT3 | ||||||||
| IRF4 | ||||||||
| EBF1 | ||||||||
| FOSL2 | ||||||||
| BATF | ||||||||
| NR3C1 | ||||||||
| RUNX3 | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr2:85764959 | rs1446668 | 2a | MAT2A | nc transcript | CTCF | CTCF | ||
| Upstream MAT2A | POLR2A | |||||||
| TAF1 | ||||||||
| RFX5 | ||||||||
| RAD21 | ||||||||
| HEY1 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| ZNF263 | ||||||||
| NR3C1 | ||||||||
| CTCF | ||||||||
| MYC | ||||||||
| AR | ||||||||
| MYBL2 | ||||||||
| TEAD4 | ||||||||
| MAZ | ||||||||
| CHD2 | ||||||||
| SMC3 | ||||||||
| TBP | ||||||||
| ZNF143 | ||||||||
| CDX2 | ||||||||
| E2F6 | ||||||||
| MAX | ||||||||
| NR3C1 | ||||||||
| SIN3A | ||||||||
| YY1 | ||||||||
| REST | ||||||||
| HMGN3 | ||||||||
| chr17:47006492 | rs12601672 | 2b | UBE2Z | Downstream UBE2Z | Zfx | POLR2A | ||
| EGR1 | ||||||||
| SPI1 | ||||||||
| ELF1 | ||||||||
| chr10:30316071 | rs3739998 | 2b | KIAA1462 | Exon 2 | RELA | CTCF | ||
| MYC | ||||||||
| PAX5 | ||||||||
| ZNF143 | ||||||||
| chr8:126476378 | rs2980856 | 2b | TRIB1 gene | Intergenic region | pax-8 | JUND | ||
| region | Downstream TRIB1 | Sox17 | POLR2A | |||||
| TFAP2C | ||||||||
| MXI1 | ||||||||
| CEBPB | ||||||||
| chr9:136154303 | rs649129 | 2b | ABO gene region | Intergenic region | IRF | NFYA | ||
| Upstream ABO | POLR2A | |||||||
| FOS | ||||||||
| IRF1 | ||||||||
| NFYB | ||||||||
| PML | ||||||||
| chr10:104840966 | rs12219901 | 2b | CNNM2 gene | Intergenic region | SRF | POLR2A | ||
| region | Downstream CXCL12 | |||||||
| chr10:44778545 | rs1746052 | 2b | CXCL12 gene | Intergenic region | GATA1 | TAL1 | ||
| region | Downstream CXCL12 | |||||||
| chr21:35593826 | rs28451064 | 2b | LINC00310 gene | Intergenic region | PPAR | SP1 | ||
| region | Downstream | FOXA2 | ||||||
| LINC00310 | ||||||||
| chr17:2098271 | rs7217687 | 2b | SMG6 | Intron 13 | - | NF-1 | SIN3A | |
| TCF12 | ||||||||
| MAX | ||||||||
| YY1 | ||||||||
| ZNF263 | ||||||||
| EP300 | ||||||||
| TEAD4 | ||||||||
| chr13:110960711 | rs4773144 | 2b | COL4A2 | Intron 3 | STAT3:STAT3 | POLR2A | ||
| EZH2 | ||||||||
| chr14:100116251 | rs28391527 | 2b | HHIPL1 | Intron 3 | MyoD | BHLHE40 | ||
| SCRT1 | USF1 | |||||||
| FIGLA | FOXA1 | |||||||
| MAX | ||||||||
| chr1:154404335 | rs7549250 | 2b | IL6R | Intron 3 | TBX15 | MXI1 | ||
| FOS | ||||||||
| JUNB | ||||||||
| MAX | ||||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| FOSL1 | ||||||||
| MAFK | ||||||||
| RCOR1 | ||||||||
| MYC | ||||||||
| USF2 | ||||||||
| TEAD4 | ||||||||
| RCOR1 | ||||||||
| YY1 | ||||||||
| chr1:154404379 | rs7549338 | 2b | IL6R | Intron 3 | GR | FOS | ||
| AR | JUNB | |||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr1:154404405 | rs7553796 | 2b | IL6R | Intron 3 | NF-kappaB, | FOS | ||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr14:100127439 | rs4624107 | 2b | HHIPL1 | Intron 7 | Pax5 | JUND | ||
| chr10:91011457 | rs1332328 | 2b | LIPA | Intron 9 | UF1H3BETA | CREBBP | ||
| ZNF263 | ||||||||
| CDX2 | ||||||||
| ELF1 | ||||||||
| ZEB1 | ||||||||
| TBP | ||||||||
| TFAPC2 | ||||||||
| TBP | ||||||||
| POLR2A | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| HEY1 | ||||||||
| chr17:2117944 | rs2281727 | 2b | SMG6 | Intron 13 | SRY | CREBBP | ||
| Srf | EP300 | |||||||
| Zfp105 | STAT3 | |||||||
| TRIM28 | ||||||||
| MYC | ||||||||
| RBBP5 | ||||||||
| chr8:126479314 | rs6982636 | 2b | LOC105375745 | Intron1 | MAF | SMARCC1 | ||
| RFX3 | ||||||||
| POLR2A | ||||||||
| GATA2 | ||||||||
| CHD2 | ||||||||
| GTF2F1 | ||||||||
| chr14:100125720 | rs7145262 | 2b | HHIPL1 | Intron4 | ESR2 | SMARC4 | ||
| ZBTB7A | ||||||||
| SMARB1 | ||||||||
| POLR2A | ||||||||
| EZH2 | ||||||||
| RAD21 | ||||||||
| BACH1 | ||||||||
| chr10:104677125 | rs12221064 | 2b | CNNM2 | Upstream CNNM2 | MAZR, | CTCF, ETS1 | ||
| ETS1 | ||||||||
| chr8:126478349 | rs2980853 | 2b | LOC105375745 | Upstream | Pit-1, | RFX3 | ||
| LOC105375745 | ||||||||
| chr15:79152421 | rs5029904 | 2b | LOC105370915 | Upstream | NeuroD | USF1 | ||
| LOC105370915 | POLR2A | |||||||
| YY1 | ||||||||
| FOXA1 | ||||||||
| E2F4 | ||||||||
| MAX | ||||||||
| TAF7 | ||||||||
| TAF1 | ||||||||
| MXI1 | ||||||||
| chr8:126478744 | rs2001844 | 2b | LOC105375745 | Upstream | HSF1 | RFX3 | ||
| LOC105375745 | ||||||||
| chr10:91011680 | rs1332327 | 2b | LIPA | 5' UTR | AP-4, | CREBBP | ||
| CDX2 | ||||||||
| ELF1 | ||||||||
| TBP | ||||||||
| SPI1 | ||||||||
| NRF1 | ||||||||
| ELF1 | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| SPI1 | ||||||||
| PAX5 | ||||||||
| SREBF1 | ||||||||
| chr17:2102452 | rs9908888 | 2b | SMG6 | Intron10 | GR | CEBPB | ||
| AR | ||||||||
| chr10:91008878 | rs2250644 | 2b | LIPA | Intron1 | Oct-1 | RUNX3 | ||
| XBP-1 | ||||||||
| MAfb | ||||||||
| Mafk | ||||||||
| MAFB | ||||||||
| MAFK | ||||||||
| NRL | ||||||||
| chr1:109817589 | rs12740374 | 2b | CELSR2 | Exon34 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| DUXA | ||||||||
| chr1:222794090 | rs17163301 | 2b | MIA3 | Intron1 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| HNF1B | ||||||||
| DUXA | ||||||||
| chr21:35644028 | rs28591415 | 2b | pseudogene | PPAR | EP300 | |||
| FOXA1 | ||||||||
| HDAC2 | ||||||||
| NFIC | ||||||||
| SP1 | ||||||||
| chr2:203713279 | rs72934715 | 2b | ICA1L | Intron2 | HMGIY | ATF2 | ||
| NFIC | ||||||||
| EBF1 | ||||||||
| EP300 | ||||||||
| NFKB1 | ||||||||
| PAX5 | ||||||||
| chr2:203775474 | rs72936852 | 2b | WDR12 | Intron1 | AR | MAFF | ||
| chr2:203926270 | rs72934512 | 2b | NBEAL1 | Intron6 | TEAD1 | TEAD4 | ||
| TEAD3 | ||||||||
| chr8:19803092 | rs3779788 | 2b | LPL | Intron1 | TGIF | CEBPB | ||
| chr10:44757106 | rs518594 | 2b | CXCL12 | Downstream intergenic | E2A | FOXM1 | ||
| NRSE | NFIC | |||||||
| NRSF | MAX | |||||||
| EBF1 | ||||||||
| TBL1XR1 | ||||||||
| chr4:57824931 | rs7687767 | 1d | CECR6 | Sox8 | FOXA1 | |||
| chr11:5759712 | rs93138 | 1f | SWAP70 | Intron8 | ||||
| rs360136 | 1f | SWAP70 | Exon13 | |||||
| chr:457798188 | rs2227901 | 1f | REST-NOA1 | Exon6 | SPI1 | |||
| chr:1567442595 | rs17293632 | 2a | SMAD3 | Intron4 | Bach1 | SIN3A | ||
| AP-1 | TCF7L2 | |||||||
| JundM2 | TFAP2A | |||||||
| Pou1f1 | TFAP2C | |||||||
| Pou3f1 | ZNF217 | |||||||
| Sox5 | POLR2A | |||||||
| JDP2 | STAT1 | |||||||
| MXI1 | ||||||||
| TAF1 | ||||||||
| E2F1 | ||||||||
| POLR2A | ||||||||
| HDAC1 | ||||||||
| TCF7L2 | ||||||||
| MYC | ||||||||
| POLR2A | ||||||||
| ESR1 | ||||||||
| EP300 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| CEBPB | ||||||||
| EP300 | ||||||||
| FOS | ||||||||
| FOSL1 | ||||||||
| GATA3 | ||||||||
| JUNB | ||||||||
| JUN | ||||||||
| MYC | ||||||||
| NR2F2 | ||||||||
| NR3C1 | ||||||||
| RAD21 | ||||||||
| RCOR1 | ||||||||
| TCF12 | ||||||||
| JUND | ||||||||
| MAFK | ||||||||
| EGR1 | ||||||||
| MAX | ||||||||
| chr15:67441996 | rs18866316 | 2b | SMAD3 | Exon4 | AIRE | ESR1 | ||
| Elf3 | NR3C1 | |||||||
| Srf | POLR2A | |||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| chr15:67448898 | rs8032739 | 2b | MTERF | Intron4 | SRF | CEBPB | ||
| FOS | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr9:22098573 | rs4977574 | 2c | CDKN2BAS1 | Intron16 | AR | AR | ||
| NR3C1 | ||||||||
| NR3C2 | ||||||||
| Ar | ||||||||
| Elk-1 | ||||||||
| c-Ets-1(p54) | ||||||||
Putative functional SNPs and corresponding motifs, eQTL and related transcription factors (Regulome DB score < 3)
| Coordinate 0-based . | SNP ID . | RegulomeDB score . | Gene/Locus . | Position . | eQTL . | Motif . | Protein-binding . | |
|---|---|---|---|---|---|---|---|---|
| chr15:91429041 | rs1894401 | 1b | FES | Intron 2 | FES | Pax5 | SPI1 | |
| USF1 | ||||||||
| POLR2A | ||||||||
| GABPA | ||||||||
| BHLHE40 | ||||||||
| CEBPB | ||||||||
| CTCF | ||||||||
| MAX | ||||||||
| RFX5 | ||||||||
| RUNX3 | ||||||||
| STAT5A | ||||||||
| chr10:91005853 | rs2246833 | 1b | LIPA | Intron 6 | LIPA | EWSR-FLI1 | CTCF | |
| znf143 | ||||||||
| chr2:85808736 | rs1009 | 1b | VAMP8 | Exon 3 | VAMP8 | CTCF | ||
| LOC388969 | HSF1 | |||||||
| chr17:47038470 | rs4794004 | 1d | GIP | Intron 4 | ATP5G1 | Gata5 | NR3C1 | |
| UBE2Z | IN3AK20 | |||||||
| CREB1 | ||||||||
| TAF1 | ||||||||
| TCF12 | ||||||||
| CTCF | ||||||||
| POLR2A | ||||||||
| USF1 | ||||||||
| FOXA1 | ||||||||
| FOXA2 | ||||||||
| RBBP5 | ||||||||
| chr10:91002926 | rs1412444 | 1d | LIPA | Intron3 | LIPA | SAP1a | ATF2 | |
| ELK1 | FOXM1 | |||||||
| ELK3 | SP1 | |||||||
| ELK4 | SPI1 | |||||||
| MECP2 | MTA3 | |||||||
| ERF | RUNX3 | |||||||
| ERG | ||||||||
| ETS1 | ||||||||
| ETV1 | ||||||||
| ETV2 | ||||||||
| ETV3 | ||||||||
| Gabpa | ||||||||
| chr10:104873760 | rs9633712 | 1e | NT5C2 | Intron 3 | UMG5 | PU1 | SP11 | |
| ELF-1 | ||||||||
| Sfpil | ||||||||
| PU.1 | ||||||||
| c-Ets-1 | ||||||||
| chr11:116648916 | rs964184 | 1f | ZPR1 | Downstream ZRP1 | TAGLN | FOXJ2 | ||
| chr1:109818529 | rs646776 | 1f | CELSR2 | Upstream CELSR2 | PSMA5 | CTCF | ||
| HEY1 | ||||||||
| REST | ||||||||
| POLR2A | ||||||||
| ZBTB7A | ||||||||
| TAF7 | ||||||||
| chr10:104616662 | rs4409766 | 1f | BORCS7-ASMT | Intron 1 | C10orf77 | Tcf3 | BACH1 | |
| USMG5 | MAFF | |||||||
| MAFK | ||||||||
| chr17:47008206 | rs4793992 | 1f | SNF8 | Intron 7 | ATP5G1 | POLR2A | ||
| UBE2Z | TEAD4 | |||||||
| chr6:161152293 | rs4252126 | 1f | PLG | Intron 11 | PLG | CTCF | ||
| RUNX3 | ||||||||
| TEAD4 | ||||||||
| RAD21 | ||||||||
| chr6:161154231 | rs4252135 | 1f | PLG | Intron 12 | PLG | CTCF | ||
| FOXA1 | ||||||||
| NFKB1 | ||||||||
| RAD21 | ||||||||
| ZNF263 | ||||||||
| SMC3 | ||||||||
| ZNF143 | ||||||||
| FOXA2 | ||||||||
| chr10:104846177 | rs11191548 | 1f | NT5C2 gene region | Downstream NT5C2 | USMG5 | TEAD1 | ||
| TEAD3 | ||||||||
| chr10:104864613 | rs11191557 | 1f | NT5C2 | Intron 5 | USMG5 | |||
| chr10:104864677 | rs11191558 | 1f | NT5C2 | Intron 5 | USMG5 | HOXC13 | ||
| Hoxa13 | ||||||||
| Hoxc13 | ||||||||
| Hoxd12 | ||||||||
| HOXA13 | ||||||||
| HOXD9 | ||||||||
| HOXC11 | ||||||||
| chr10:104871203 | rs12413046 | 1f | NT5C2 | Intron 3 | USMG5 | NR3C1 | ||
| TRIM28 | ||||||||
| CTCF | ||||||||
| ATF2 | ||||||||
| IKZF1 | ||||||||
| TCF7L2 | ||||||||
| ZNF263 | ||||||||
| chr10:104871278 | rs10883832 | 1f | NT5C2 | Intron 3 | USMG5 | TRIM28 | ||
| TCF7L2 | ||||||||
| chr10:104913652 | rs11191582 | 1f | NT5C2 | Intron 2 | USMG5 | EP300 | ||
| NFIC | ||||||||
| TCF12 | ||||||||
| TEAD4 | ||||||||
| STAT1 | ||||||||
| ARID3A | ||||||||
| EP300 | ||||||||
| JUN | ||||||||
| RCOR1 | ||||||||
| chr10:104906210 | rs11191580 | 1f | NT5C2 | Intron 2 | USMG5 | TRIM28 | ||
| SETDB1 | ||||||||
| GATA1 | ||||||||
| GTF2F1 | ||||||||
| CEBPB | ||||||||
| FOS | ||||||||
| JUND | ||||||||
| ZNF263 | ||||||||
| chr10:104856161 | rs12412038 | 1f | NT5C2 | Intron 10 | USMG5 | Irx-3 | ||
| Irx-2 | ||||||||
| Irx-4 | ||||||||
| Irx-6 | ||||||||
| chr2:85807081 | rs1348818 | 1f | VAMP8 | Intron 2 | GGCX | HMGIY | EBF1, | |
| Mtf1 | ||||||||
| Srf | ||||||||
| Zfp105 | ||||||||
| HMGIY | ||||||||
| chr2:85805366 | rs3770098 | 1f | VAMP8 | Intron1 | VAMP8 | POLR2A | ||
| LOC388969 | BHLHE40 | |||||||
| E2F6 | ||||||||
| KDM5B | ||||||||
| MAX | ||||||||
| MXI1 | ||||||||
| MYC | ||||||||
| NFIC | ||||||||
| WRNIP1 | ||||||||
| chr2:85803541 | rs6757263 | 1f | VAMP8 | Upstream VAMP8 | GGCX | SP1 | ||
| VAMP8 | EP300 | |||||||
| LOC388969 | NFIC | |||||||
| chr17:46988596 | rs46522 | 1f | UBE2Z | Intron 2 | ATP5G1 | NFKB | ||
| UBE2Z | NFYB | |||||||
| RUNX3 | ||||||||
| chr19:45395618 | rs2075650 | 1f | TOMM40 | Intron 2 | TOMM40 | RREB1 | ||
| chr1:56996190 | rs4634932 | 1f | PLPP3 | Intron 2 | PPAP2B | POLR2A | ||
| chr2:203880833 | rs4675310 | 1f | NBEAL1 | Intron 1 | ALS2CR13 | |||
| chr10:104681142 | rs17115213 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104721125 | rs10883808 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104723619 | rs11191479 | 1f | CNNM2 | Intron 1 | USMG5 | GATA1 | ||
| TAL1 | ||||||||
| CEBPB | ||||||||
| chr10:104773363 | rs11191514 | 1f | CNNM2 | Intron 1 | USMG5 | PAX5 | ||
| chr10:104776526 | rs11191515 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104825664 | rs3781285 | 1f | CNNM2 | Intron 4 | USMG5 | NF-kappaB | IKZF1 | |
| P50:50 | ||||||||
| chr10:104835918 | rs943037 | 1f | CNNM2 | Exon 7 | USMG5 | TBX20 | ||
| Foxj1 | ||||||||
| chr8:19813701 | rs271 | 1f | LPL | Intron 6 | LPL | |||
| chr17:47039131 | rs2291725 | 1f | GIP | Exon 4 | GIP | GATA2 | ||
| TCF4 | ||||||||
| FOSL2 | ||||||||
| EGR1 | ||||||||
| ELF1 | ||||||||
| FOS | ||||||||
| NR3C1 | ||||||||
| EP300 | ||||||||
| RXRA | ||||||||
| CHD2 | ||||||||
| JUND | ||||||||
| POLR2A | ||||||||
| RAD21 | ||||||||
| FOSL1 | ||||||||
| REST | ||||||||
| chr2:203880991 | rs2351524 | 1f | NBEAL1 | 5' UTR | ALS2CR13 | |||
| chr1:109818305 | rs629301 | 1f | CELSR2 | 3' UTR | PSRC1 | CTCF | ||
| POLR2A | ||||||||
| chr2:85774675 | rs6547621 | 1f | 3' UTR | GGCX | ELK4 | |||
| POLR2A | ||||||||
| chr10:104660003 | rs11191454 | 1f | AS3MT | Intron10 | USMG5 | |||
| chr10:104685298 | rs12411886 | 1f | CNNM2 | Intron1 | USMG5 | Zec | ||
| chr10:104719095 | rs12413409 | 1f | CNNM2 | Intron1 | USMG5 | POLR3A | ||
| chr10:104764270 | rs11191499 | 1f | CNNM2 | Intron1 | USMG5 | |||
| chr17:47014126 | rs1994970 | 1f | SNF8 | Intron4 | ATP5G1 | TFII-I | ||
| UBE2Z | ||||||||
| chr2:85742296 | rs2886722 | 1f | Pseudogene | LOC388969 | TCF7L2 | |||
| chr2:85783127 | rs6738645 | 1f | GGCX | Intron5 | Evi-1 | POLR2A | ||
| chr2:85794296 | rs10187424 | 1f | Pseudogene | GGCX | ||||
| LOC388969 | ||||||||
| chr5:131723064 | rs17689550 | 1f | RAPGEF6 | |||||
| chr7:129663495 | rs11556924 | 1f | ZC3HC1 | Exon8 | KIAA0265 | |||
| chr17:4702833 | rs11079844 | 1f | Pseudogene | ATP5G1 | ||||
| chr17: 47005192 | rs15563 | 1f | UBE2Z | Exon7 | ATP5G1 | PRDM1 | ||
| chr17:47047113 | rs3848460 | 1f | UBE2Z | ATP5G1 | CEBPB | |||
| chr10:104680136 | rs2297787 | 2a | CNNM2 | Intron 1 | Freac-7 | FOXA1 | ||
| HFH3(FOXI1) | SIN3A | |||||||
| HNF3alpha | ZNF263 | |||||||
| FOXP1 | HNF4G | |||||||
| Elf3 | FOXA1 | |||||||
| Foxl1 | ||||||||
| Srf | ||||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| Zfp105 | ||||||||
| HFH(FOXl1) | ||||||||
| chr1:56948289 | rs72664304 | 2a | C8B | Intron 6 | FOXA1 | FOXA1 | ||
| Foxa2 | FOXA2 | |||||||
| TCF4 | ||||||||
| SP1 | ||||||||
| HNF4G | ||||||||
| HNF4A | ||||||||
| HDAC2 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| chr17:46993232 | rs3744608 | 2a | UBE2Z | Intron 3 | Zfp740 | SPI1 | ||
| MZF1 | POLR2A | |||||||
| MAZR | IKZF1 | |||||||
| SP1 | MAX | |||||||
| SP1:SP3 | TFAP2A | |||||||
| WT1ZNF21 | TFAP2C | |||||||
| Zfp281 | SP1 | |||||||
| ZFp740 | CEBPB | |||||||
| WT1 | NR3C1 | |||||||
| ZNF219 | BATF | |||||||
| ZNF740 | BCL11A | |||||||
| SP4 | MEF2A | |||||||
| NFKB1 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| STAT3 | ||||||||
| IRF4 | ||||||||
| EBF1 | ||||||||
| FOSL2 | ||||||||
| BATF | ||||||||
| NR3C1 | ||||||||
| RUNX3 | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr2:85764959 | rs1446668 | 2a | MAT2A | nc transcript | CTCF | CTCF | ||
| Upstream MAT2A | POLR2A | |||||||
| TAF1 | ||||||||
| RFX5 | ||||||||
| RAD21 | ||||||||
| HEY1 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| ZNF263 | ||||||||
| NR3C1 | ||||||||
| CTCF | ||||||||
| MYC | ||||||||
| AR | ||||||||
| MYBL2 | ||||||||
| TEAD4 | ||||||||
| MAZ | ||||||||
| CHD2 | ||||||||
| SMC3 | ||||||||
| TBP | ||||||||
| ZNF143 | ||||||||
| CDX2 | ||||||||
| E2F6 | ||||||||
| MAX | ||||||||
| NR3C1 | ||||||||
| SIN3A | ||||||||
| YY1 | ||||||||
| REST | ||||||||
| HMGN3 | ||||||||
| chr17:47006492 | rs12601672 | 2b | UBE2Z | Downstream UBE2Z | Zfx | POLR2A | ||
| EGR1 | ||||||||
| SPI1 | ||||||||
| ELF1 | ||||||||
| chr10:30316071 | rs3739998 | 2b | KIAA1462 | Exon 2 | RELA | CTCF | ||
| MYC | ||||||||
| PAX5 | ||||||||
| ZNF143 | ||||||||
| chr8:126476378 | rs2980856 | 2b | TRIB1 gene | Intergenic region | pax-8 | JUND | ||
| region | Downstream TRIB1 | Sox17 | POLR2A | |||||
| TFAP2C | ||||||||
| MXI1 | ||||||||
| CEBPB | ||||||||
| chr9:136154303 | rs649129 | 2b | ABO gene region | Intergenic region | IRF | NFYA | ||
| Upstream ABO | POLR2A | |||||||
| FOS | ||||||||
| IRF1 | ||||||||
| NFYB | ||||||||
| PML | ||||||||
| chr10:104840966 | rs12219901 | 2b | CNNM2 gene | Intergenic region | SRF | POLR2A | ||
| region | Downstream CXCL12 | |||||||
| chr10:44778545 | rs1746052 | 2b | CXCL12 gene | Intergenic region | GATA1 | TAL1 | ||
| region | Downstream CXCL12 | |||||||
| chr21:35593826 | rs28451064 | 2b | LINC00310 gene | Intergenic region | PPAR | SP1 | ||
| region | Downstream | FOXA2 | ||||||
| LINC00310 | ||||||||
| chr17:2098271 | rs7217687 | 2b | SMG6 | Intron 13 | - | NF-1 | SIN3A | |
| TCF12 | ||||||||
| MAX | ||||||||
| YY1 | ||||||||
| ZNF263 | ||||||||
| EP300 | ||||||||
| TEAD4 | ||||||||
| chr13:110960711 | rs4773144 | 2b | COL4A2 | Intron 3 | STAT3:STAT3 | POLR2A | ||
| EZH2 | ||||||||
| chr14:100116251 | rs28391527 | 2b | HHIPL1 | Intron 3 | MyoD | BHLHE40 | ||
| SCRT1 | USF1 | |||||||
| FIGLA | FOXA1 | |||||||
| MAX | ||||||||
| chr1:154404335 | rs7549250 | 2b | IL6R | Intron 3 | TBX15 | MXI1 | ||
| FOS | ||||||||
| JUNB | ||||||||
| MAX | ||||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| FOSL1 | ||||||||
| MAFK | ||||||||
| RCOR1 | ||||||||
| MYC | ||||||||
| USF2 | ||||||||
| TEAD4 | ||||||||
| RCOR1 | ||||||||
| YY1 | ||||||||
| chr1:154404379 | rs7549338 | 2b | IL6R | Intron 3 | GR | FOS | ||
| AR | JUNB | |||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr1:154404405 | rs7553796 | 2b | IL6R | Intron 3 | NF-kappaB, | FOS | ||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr14:100127439 | rs4624107 | 2b | HHIPL1 | Intron 7 | Pax5 | JUND | ||
| chr10:91011457 | rs1332328 | 2b | LIPA | Intron 9 | UF1H3BETA | CREBBP | ||
| ZNF263 | ||||||||
| CDX2 | ||||||||
| ELF1 | ||||||||
| ZEB1 | ||||||||
| TBP | ||||||||
| TFAPC2 | ||||||||
| TBP | ||||||||
| POLR2A | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| HEY1 | ||||||||
| chr17:2117944 | rs2281727 | 2b | SMG6 | Intron 13 | SRY | CREBBP | ||
| Srf | EP300 | |||||||
| Zfp105 | STAT3 | |||||||
| TRIM28 | ||||||||
| MYC | ||||||||
| RBBP5 | ||||||||
| chr8:126479314 | rs6982636 | 2b | LOC105375745 | Intron1 | MAF | SMARCC1 | ||
| RFX3 | ||||||||
| POLR2A | ||||||||
| GATA2 | ||||||||
| CHD2 | ||||||||
| GTF2F1 | ||||||||
| chr14:100125720 | rs7145262 | 2b | HHIPL1 | Intron4 | ESR2 | SMARC4 | ||
| ZBTB7A | ||||||||
| SMARB1 | ||||||||
| POLR2A | ||||||||
| EZH2 | ||||||||
| RAD21 | ||||||||
| BACH1 | ||||||||
| chr10:104677125 | rs12221064 | 2b | CNNM2 | Upstream CNNM2 | MAZR, | CTCF, ETS1 | ||
| ETS1 | ||||||||
| chr8:126478349 | rs2980853 | 2b | LOC105375745 | Upstream | Pit-1, | RFX3 | ||
| LOC105375745 | ||||||||
| chr15:79152421 | rs5029904 | 2b | LOC105370915 | Upstream | NeuroD | USF1 | ||
| LOC105370915 | POLR2A | |||||||
| YY1 | ||||||||
| FOXA1 | ||||||||
| E2F4 | ||||||||
| MAX | ||||||||
| TAF7 | ||||||||
| TAF1 | ||||||||
| MXI1 | ||||||||
| chr8:126478744 | rs2001844 | 2b | LOC105375745 | Upstream | HSF1 | RFX3 | ||
| LOC105375745 | ||||||||
| chr10:91011680 | rs1332327 | 2b | LIPA | 5' UTR | AP-4, | CREBBP | ||
| CDX2 | ||||||||
| ELF1 | ||||||||
| TBP | ||||||||
| SPI1 | ||||||||
| NRF1 | ||||||||
| ELF1 | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| SPI1 | ||||||||
| PAX5 | ||||||||
| SREBF1 | ||||||||
| chr17:2102452 | rs9908888 | 2b | SMG6 | Intron10 | GR | CEBPB | ||
| AR | ||||||||
| chr10:91008878 | rs2250644 | 2b | LIPA | Intron1 | Oct-1 | RUNX3 | ||
| XBP-1 | ||||||||
| MAfb | ||||||||
| Mafk | ||||||||
| MAFB | ||||||||
| MAFK | ||||||||
| NRL | ||||||||
| chr1:109817589 | rs12740374 | 2b | CELSR2 | Exon34 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| DUXA | ||||||||
| chr1:222794090 | rs17163301 | 2b | MIA3 | Intron1 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| HNF1B | ||||||||
| DUXA | ||||||||
| chr21:35644028 | rs28591415 | 2b | pseudogene | PPAR | EP300 | |||
| FOXA1 | ||||||||
| HDAC2 | ||||||||
| NFIC | ||||||||
| SP1 | ||||||||
| chr2:203713279 | rs72934715 | 2b | ICA1L | Intron2 | HMGIY | ATF2 | ||
| NFIC | ||||||||
| EBF1 | ||||||||
| EP300 | ||||||||
| NFKB1 | ||||||||
| PAX5 | ||||||||
| chr2:203775474 | rs72936852 | 2b | WDR12 | Intron1 | AR | MAFF | ||
| chr2:203926270 | rs72934512 | 2b | NBEAL1 | Intron6 | TEAD1 | TEAD4 | ||
| TEAD3 | ||||||||
| chr8:19803092 | rs3779788 | 2b | LPL | Intron1 | TGIF | CEBPB | ||
| chr10:44757106 | rs518594 | 2b | CXCL12 | Downstream intergenic | E2A | FOXM1 | ||
| NRSE | NFIC | |||||||
| NRSF | MAX | |||||||
| EBF1 | ||||||||
| TBL1XR1 | ||||||||
| chr4:57824931 | rs7687767 | 1d | CECR6 | Sox8 | FOXA1 | |||
| chr11:5759712 | rs93138 | 1f | SWAP70 | Intron8 | ||||
| rs360136 | 1f | SWAP70 | Exon13 | |||||
| chr:457798188 | rs2227901 | 1f | REST-NOA1 | Exon6 | SPI1 | |||
| chr:1567442595 | rs17293632 | 2a | SMAD3 | Intron4 | Bach1 | SIN3A | ||
| AP-1 | TCF7L2 | |||||||
| JundM2 | TFAP2A | |||||||
| Pou1f1 | TFAP2C | |||||||
| Pou3f1 | ZNF217 | |||||||
| Sox5 | POLR2A | |||||||
| JDP2 | STAT1 | |||||||
| MXI1 | ||||||||
| TAF1 | ||||||||
| E2F1 | ||||||||
| POLR2A | ||||||||
| HDAC1 | ||||||||
| TCF7L2 | ||||||||
| MYC | ||||||||
| POLR2A | ||||||||
| ESR1 | ||||||||
| EP300 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| CEBPB | ||||||||
| EP300 | ||||||||
| FOS | ||||||||
| FOSL1 | ||||||||
| GATA3 | ||||||||
| JUNB | ||||||||
| JUN | ||||||||
| MYC | ||||||||
| NR2F2 | ||||||||
| NR3C1 | ||||||||
| RAD21 | ||||||||
| RCOR1 | ||||||||
| TCF12 | ||||||||
| JUND | ||||||||
| MAFK | ||||||||
| EGR1 | ||||||||
| MAX | ||||||||
| chr15:67441996 | rs18866316 | 2b | SMAD3 | Exon4 | AIRE | ESR1 | ||
| Elf3 | NR3C1 | |||||||
| Srf | POLR2A | |||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| chr15:67448898 | rs8032739 | 2b | MTERF | Intron4 | SRF | CEBPB | ||
| FOS | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr9:22098573 | rs4977574 | 2c | CDKN2BAS1 | Intron16 | AR | AR | ||
| NR3C1 | ||||||||
| NR3C2 | ||||||||
| Ar | ||||||||
| Elk-1 | ||||||||
| c-Ets-1(p54) | ||||||||
| Coordinate 0-based . | SNP ID . | RegulomeDB score . | Gene/Locus . | Position . | eQTL . | Motif . | Protein-binding . | |
|---|---|---|---|---|---|---|---|---|
| chr15:91429041 | rs1894401 | 1b | FES | Intron 2 | FES | Pax5 | SPI1 | |
| USF1 | ||||||||
| POLR2A | ||||||||
| GABPA | ||||||||
| BHLHE40 | ||||||||
| CEBPB | ||||||||
| CTCF | ||||||||
| MAX | ||||||||
| RFX5 | ||||||||
| RUNX3 | ||||||||
| STAT5A | ||||||||
| chr10:91005853 | rs2246833 | 1b | LIPA | Intron 6 | LIPA | EWSR-FLI1 | CTCF | |
| znf143 | ||||||||
| chr2:85808736 | rs1009 | 1b | VAMP8 | Exon 3 | VAMP8 | CTCF | ||
| LOC388969 | HSF1 | |||||||
| chr17:47038470 | rs4794004 | 1d | GIP | Intron 4 | ATP5G1 | Gata5 | NR3C1 | |
| UBE2Z | IN3AK20 | |||||||
| CREB1 | ||||||||
| TAF1 | ||||||||
| TCF12 | ||||||||
| CTCF | ||||||||
| POLR2A | ||||||||
| USF1 | ||||||||
| FOXA1 | ||||||||
| FOXA2 | ||||||||
| RBBP5 | ||||||||
| chr10:91002926 | rs1412444 | 1d | LIPA | Intron3 | LIPA | SAP1a | ATF2 | |
| ELK1 | FOXM1 | |||||||
| ELK3 | SP1 | |||||||
| ELK4 | SPI1 | |||||||
| MECP2 | MTA3 | |||||||
| ERF | RUNX3 | |||||||
| ERG | ||||||||
| ETS1 | ||||||||
| ETV1 | ||||||||
| ETV2 | ||||||||
| ETV3 | ||||||||
| Gabpa | ||||||||
| chr10:104873760 | rs9633712 | 1e | NT5C2 | Intron 3 | UMG5 | PU1 | SP11 | |
| ELF-1 | ||||||||
| Sfpil | ||||||||
| PU.1 | ||||||||
| c-Ets-1 | ||||||||
| chr11:116648916 | rs964184 | 1f | ZPR1 | Downstream ZRP1 | TAGLN | FOXJ2 | ||
| chr1:109818529 | rs646776 | 1f | CELSR2 | Upstream CELSR2 | PSMA5 | CTCF | ||
| HEY1 | ||||||||
| REST | ||||||||
| POLR2A | ||||||||
| ZBTB7A | ||||||||
| TAF7 | ||||||||
| chr10:104616662 | rs4409766 | 1f | BORCS7-ASMT | Intron 1 | C10orf77 | Tcf3 | BACH1 | |
| USMG5 | MAFF | |||||||
| MAFK | ||||||||
| chr17:47008206 | rs4793992 | 1f | SNF8 | Intron 7 | ATP5G1 | POLR2A | ||
| UBE2Z | TEAD4 | |||||||
| chr6:161152293 | rs4252126 | 1f | PLG | Intron 11 | PLG | CTCF | ||
| RUNX3 | ||||||||
| TEAD4 | ||||||||
| RAD21 | ||||||||
| chr6:161154231 | rs4252135 | 1f | PLG | Intron 12 | PLG | CTCF | ||
| FOXA1 | ||||||||
| NFKB1 | ||||||||
| RAD21 | ||||||||
| ZNF263 | ||||||||
| SMC3 | ||||||||
| ZNF143 | ||||||||
| FOXA2 | ||||||||
| chr10:104846177 | rs11191548 | 1f | NT5C2 gene region | Downstream NT5C2 | USMG5 | TEAD1 | ||
| TEAD3 | ||||||||
| chr10:104864613 | rs11191557 | 1f | NT5C2 | Intron 5 | USMG5 | |||
| chr10:104864677 | rs11191558 | 1f | NT5C2 | Intron 5 | USMG5 | HOXC13 | ||
| Hoxa13 | ||||||||
| Hoxc13 | ||||||||
| Hoxd12 | ||||||||
| HOXA13 | ||||||||
| HOXD9 | ||||||||
| HOXC11 | ||||||||
| chr10:104871203 | rs12413046 | 1f | NT5C2 | Intron 3 | USMG5 | NR3C1 | ||
| TRIM28 | ||||||||
| CTCF | ||||||||
| ATF2 | ||||||||
| IKZF1 | ||||||||
| TCF7L2 | ||||||||
| ZNF263 | ||||||||
| chr10:104871278 | rs10883832 | 1f | NT5C2 | Intron 3 | USMG5 | TRIM28 | ||
| TCF7L2 | ||||||||
| chr10:104913652 | rs11191582 | 1f | NT5C2 | Intron 2 | USMG5 | EP300 | ||
| NFIC | ||||||||
| TCF12 | ||||||||
| TEAD4 | ||||||||
| STAT1 | ||||||||
| ARID3A | ||||||||
| EP300 | ||||||||
| JUN | ||||||||
| RCOR1 | ||||||||
| chr10:104906210 | rs11191580 | 1f | NT5C2 | Intron 2 | USMG5 | TRIM28 | ||
| SETDB1 | ||||||||
| GATA1 | ||||||||
| GTF2F1 | ||||||||
| CEBPB | ||||||||
| FOS | ||||||||
| JUND | ||||||||
| ZNF263 | ||||||||
| chr10:104856161 | rs12412038 | 1f | NT5C2 | Intron 10 | USMG5 | Irx-3 | ||
| Irx-2 | ||||||||
| Irx-4 | ||||||||
| Irx-6 | ||||||||
| chr2:85807081 | rs1348818 | 1f | VAMP8 | Intron 2 | GGCX | HMGIY | EBF1, | |
| Mtf1 | ||||||||
| Srf | ||||||||
| Zfp105 | ||||||||
| HMGIY | ||||||||
| chr2:85805366 | rs3770098 | 1f | VAMP8 | Intron1 | VAMP8 | POLR2A | ||
| LOC388969 | BHLHE40 | |||||||
| E2F6 | ||||||||
| KDM5B | ||||||||
| MAX | ||||||||
| MXI1 | ||||||||
| MYC | ||||||||
| NFIC | ||||||||
| WRNIP1 | ||||||||
| chr2:85803541 | rs6757263 | 1f | VAMP8 | Upstream VAMP8 | GGCX | SP1 | ||
| VAMP8 | EP300 | |||||||
| LOC388969 | NFIC | |||||||
| chr17:46988596 | rs46522 | 1f | UBE2Z | Intron 2 | ATP5G1 | NFKB | ||
| UBE2Z | NFYB | |||||||
| RUNX3 | ||||||||
| chr19:45395618 | rs2075650 | 1f | TOMM40 | Intron 2 | TOMM40 | RREB1 | ||
| chr1:56996190 | rs4634932 | 1f | PLPP3 | Intron 2 | PPAP2B | POLR2A | ||
| chr2:203880833 | rs4675310 | 1f | NBEAL1 | Intron 1 | ALS2CR13 | |||
| chr10:104681142 | rs17115213 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104721125 | rs10883808 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104723619 | rs11191479 | 1f | CNNM2 | Intron 1 | USMG5 | GATA1 | ||
| TAL1 | ||||||||
| CEBPB | ||||||||
| chr10:104773363 | rs11191514 | 1f | CNNM2 | Intron 1 | USMG5 | PAX5 | ||
| chr10:104776526 | rs11191515 | 1f | CNNM2 | Intron 1 | USMG5 | |||
| chr10:104825664 | rs3781285 | 1f | CNNM2 | Intron 4 | USMG5 | NF-kappaB | IKZF1 | |
| P50:50 | ||||||||
| chr10:104835918 | rs943037 | 1f | CNNM2 | Exon 7 | USMG5 | TBX20 | ||
| Foxj1 | ||||||||
| chr8:19813701 | rs271 | 1f | LPL | Intron 6 | LPL | |||
| chr17:47039131 | rs2291725 | 1f | GIP | Exon 4 | GIP | GATA2 | ||
| TCF4 | ||||||||
| FOSL2 | ||||||||
| EGR1 | ||||||||
| ELF1 | ||||||||
| FOS | ||||||||
| NR3C1 | ||||||||
| EP300 | ||||||||
| RXRA | ||||||||
| CHD2 | ||||||||
| JUND | ||||||||
| POLR2A | ||||||||
| RAD21 | ||||||||
| FOSL1 | ||||||||
| REST | ||||||||
| chr2:203880991 | rs2351524 | 1f | NBEAL1 | 5' UTR | ALS2CR13 | |||
| chr1:109818305 | rs629301 | 1f | CELSR2 | 3' UTR | PSRC1 | CTCF | ||
| POLR2A | ||||||||
| chr2:85774675 | rs6547621 | 1f | 3' UTR | GGCX | ELK4 | |||
| POLR2A | ||||||||
| chr10:104660003 | rs11191454 | 1f | AS3MT | Intron10 | USMG5 | |||
| chr10:104685298 | rs12411886 | 1f | CNNM2 | Intron1 | USMG5 | Zec | ||
| chr10:104719095 | rs12413409 | 1f | CNNM2 | Intron1 | USMG5 | POLR3A | ||
| chr10:104764270 | rs11191499 | 1f | CNNM2 | Intron1 | USMG5 | |||
| chr17:47014126 | rs1994970 | 1f | SNF8 | Intron4 | ATP5G1 | TFII-I | ||
| UBE2Z | ||||||||
| chr2:85742296 | rs2886722 | 1f | Pseudogene | LOC388969 | TCF7L2 | |||
| chr2:85783127 | rs6738645 | 1f | GGCX | Intron5 | Evi-1 | POLR2A | ||
| chr2:85794296 | rs10187424 | 1f | Pseudogene | GGCX | ||||
| LOC388969 | ||||||||
| chr5:131723064 | rs17689550 | 1f | RAPGEF6 | |||||
| chr7:129663495 | rs11556924 | 1f | ZC3HC1 | Exon8 | KIAA0265 | |||
| chr17:4702833 | rs11079844 | 1f | Pseudogene | ATP5G1 | ||||
| chr17: 47005192 | rs15563 | 1f | UBE2Z | Exon7 | ATP5G1 | PRDM1 | ||
| chr17:47047113 | rs3848460 | 1f | UBE2Z | ATP5G1 | CEBPB | |||
| chr10:104680136 | rs2297787 | 2a | CNNM2 | Intron 1 | Freac-7 | FOXA1 | ||
| HFH3(FOXI1) | SIN3A | |||||||
| HNF3alpha | ZNF263 | |||||||
| FOXP1 | HNF4G | |||||||
| Elf3 | FOXA1 | |||||||
| Foxl1 | ||||||||
| Srf | ||||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| Zfp105 | ||||||||
| HFH(FOXl1) | ||||||||
| chr1:56948289 | rs72664304 | 2a | C8B | Intron 6 | FOXA1 | FOXA1 | ||
| Foxa2 | FOXA2 | |||||||
| TCF4 | ||||||||
| SP1 | ||||||||
| HNF4G | ||||||||
| HNF4A | ||||||||
| HDAC2 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| chr17:46993232 | rs3744608 | 2a | UBE2Z | Intron 3 | Zfp740 | SPI1 | ||
| MZF1 | POLR2A | |||||||
| MAZR | IKZF1 | |||||||
| SP1 | MAX | |||||||
| SP1:SP3 | TFAP2A | |||||||
| WT1ZNF21 | TFAP2C | |||||||
| Zfp281 | SP1 | |||||||
| ZFp740 | CEBPB | |||||||
| WT1 | NR3C1 | |||||||
| ZNF219 | BATF | |||||||
| ZNF740 | BCL11A | |||||||
| SP4 | MEF2A | |||||||
| NFKB1 | ||||||||
| JUND | ||||||||
| EP300 | ||||||||
| STAT3 | ||||||||
| IRF4 | ||||||||
| EBF1 | ||||||||
| FOSL2 | ||||||||
| BATF | ||||||||
| NR3C1 | ||||||||
| RUNX3 | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr2:85764959 | rs1446668 | 2a | MAT2A | nc transcript | CTCF | CTCF | ||
| Upstream MAT2A | POLR2A | |||||||
| TAF1 | ||||||||
| RFX5 | ||||||||
| RAD21 | ||||||||
| HEY1 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| ZNF263 | ||||||||
| NR3C1 | ||||||||
| CTCF | ||||||||
| MYC | ||||||||
| AR | ||||||||
| MYBL2 | ||||||||
| TEAD4 | ||||||||
| MAZ | ||||||||
| CHD2 | ||||||||
| SMC3 | ||||||||
| TBP | ||||||||
| ZNF143 | ||||||||
| CDX2 | ||||||||
| E2F6 | ||||||||
| MAX | ||||||||
| NR3C1 | ||||||||
| SIN3A | ||||||||
| YY1 | ||||||||
| REST | ||||||||
| HMGN3 | ||||||||
| chr17:47006492 | rs12601672 | 2b | UBE2Z | Downstream UBE2Z | Zfx | POLR2A | ||
| EGR1 | ||||||||
| SPI1 | ||||||||
| ELF1 | ||||||||
| chr10:30316071 | rs3739998 | 2b | KIAA1462 | Exon 2 | RELA | CTCF | ||
| MYC | ||||||||
| PAX5 | ||||||||
| ZNF143 | ||||||||
| chr8:126476378 | rs2980856 | 2b | TRIB1 gene | Intergenic region | pax-8 | JUND | ||
| region | Downstream TRIB1 | Sox17 | POLR2A | |||||
| TFAP2C | ||||||||
| MXI1 | ||||||||
| CEBPB | ||||||||
| chr9:136154303 | rs649129 | 2b | ABO gene region | Intergenic region | IRF | NFYA | ||
| Upstream ABO | POLR2A | |||||||
| FOS | ||||||||
| IRF1 | ||||||||
| NFYB | ||||||||
| PML | ||||||||
| chr10:104840966 | rs12219901 | 2b | CNNM2 gene | Intergenic region | SRF | POLR2A | ||
| region | Downstream CXCL12 | |||||||
| chr10:44778545 | rs1746052 | 2b | CXCL12 gene | Intergenic region | GATA1 | TAL1 | ||
| region | Downstream CXCL12 | |||||||
| chr21:35593826 | rs28451064 | 2b | LINC00310 gene | Intergenic region | PPAR | SP1 | ||
| region | Downstream | FOXA2 | ||||||
| LINC00310 | ||||||||
| chr17:2098271 | rs7217687 | 2b | SMG6 | Intron 13 | - | NF-1 | SIN3A | |
| TCF12 | ||||||||
| MAX | ||||||||
| YY1 | ||||||||
| ZNF263 | ||||||||
| EP300 | ||||||||
| TEAD4 | ||||||||
| chr13:110960711 | rs4773144 | 2b | COL4A2 | Intron 3 | STAT3:STAT3 | POLR2A | ||
| EZH2 | ||||||||
| chr14:100116251 | rs28391527 | 2b | HHIPL1 | Intron 3 | MyoD | BHLHE40 | ||
| SCRT1 | USF1 | |||||||
| FIGLA | FOXA1 | |||||||
| MAX | ||||||||
| chr1:154404335 | rs7549250 | 2b | IL6R | Intron 3 | TBX15 | MXI1 | ||
| FOS | ||||||||
| JUNB | ||||||||
| MAX | ||||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| FOSL1 | ||||||||
| MAFK | ||||||||
| RCOR1 | ||||||||
| MYC | ||||||||
| USF2 | ||||||||
| TEAD4 | ||||||||
| RCOR1 | ||||||||
| YY1 | ||||||||
| chr1:154404379 | rs7549338 | 2b | IL6R | Intron 3 | GR | FOS | ||
| AR | JUNB | |||||||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr1:154404405 | rs7553796 | 2b | IL6R | Intron 3 | NF-kappaB, | FOS | ||
| JUND | ||||||||
| JUN | ||||||||
| STAT3 | ||||||||
| chr14:100127439 | rs4624107 | 2b | HHIPL1 | Intron 7 | Pax5 | JUND | ||
| chr10:91011457 | rs1332328 | 2b | LIPA | Intron 9 | UF1H3BETA | CREBBP | ||
| ZNF263 | ||||||||
| CDX2 | ||||||||
| ELF1 | ||||||||
| ZEB1 | ||||||||
| TBP | ||||||||
| TFAPC2 | ||||||||
| TBP | ||||||||
| POLR2A | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| HEY1 | ||||||||
| chr17:2117944 | rs2281727 | 2b | SMG6 | Intron 13 | SRY | CREBBP | ||
| Srf | EP300 | |||||||
| Zfp105 | STAT3 | |||||||
| TRIM28 | ||||||||
| MYC | ||||||||
| RBBP5 | ||||||||
| chr8:126479314 | rs6982636 | 2b | LOC105375745 | Intron1 | MAF | SMARCC1 | ||
| RFX3 | ||||||||
| POLR2A | ||||||||
| GATA2 | ||||||||
| CHD2 | ||||||||
| GTF2F1 | ||||||||
| chr14:100125720 | rs7145262 | 2b | HHIPL1 | Intron4 | ESR2 | SMARC4 | ||
| ZBTB7A | ||||||||
| SMARB1 | ||||||||
| POLR2A | ||||||||
| EZH2 | ||||||||
| RAD21 | ||||||||
| BACH1 | ||||||||
| chr10:104677125 | rs12221064 | 2b | CNNM2 | Upstream CNNM2 | MAZR, | CTCF, ETS1 | ||
| ETS1 | ||||||||
| chr8:126478349 | rs2980853 | 2b | LOC105375745 | Upstream | Pit-1, | RFX3 | ||
| LOC105375745 | ||||||||
| chr15:79152421 | rs5029904 | 2b | LOC105370915 | Upstream | NeuroD | USF1 | ||
| LOC105370915 | POLR2A | |||||||
| YY1 | ||||||||
| FOXA1 | ||||||||
| E2F4 | ||||||||
| MAX | ||||||||
| TAF7 | ||||||||
| TAF1 | ||||||||
| MXI1 | ||||||||
| chr8:126478744 | rs2001844 | 2b | LOC105375745 | Upstream | HSF1 | RFX3 | ||
| LOC105375745 | ||||||||
| chr10:91011680 | rs1332327 | 2b | LIPA | 5' UTR | AP-4, | CREBBP | ||
| CDX2 | ||||||||
| ELF1 | ||||||||
| TBP | ||||||||
| SPI1 | ||||||||
| NRF1 | ||||||||
| ELF1 | ||||||||
| ETS1 | ||||||||
| GABPA | ||||||||
| SPI1 | ||||||||
| PAX5 | ||||||||
| SREBF1 | ||||||||
| chr17:2102452 | rs9908888 | 2b | SMG6 | Intron10 | GR | CEBPB | ||
| AR | ||||||||
| chr10:91008878 | rs2250644 | 2b | LIPA | Intron1 | Oct-1 | RUNX3 | ||
| XBP-1 | ||||||||
| MAfb | ||||||||
| Mafk | ||||||||
| MAFB | ||||||||
| MAFK | ||||||||
| NRL | ||||||||
| chr1:109817589 | rs12740374 | 2b | CELSR2 | Exon34 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| DUXA | ||||||||
| chr1:222794090 | rs17163301 | 2b | MIA3 | Intron1 | HNF1 | EBF1 | ||
| HNF1A | ||||||||
| HNF1B | ||||||||
| DUXA | ||||||||
| chr21:35644028 | rs28591415 | 2b | pseudogene | PPAR | EP300 | |||
| FOXA1 | ||||||||
| HDAC2 | ||||||||
| NFIC | ||||||||
| SP1 | ||||||||
| chr2:203713279 | rs72934715 | 2b | ICA1L | Intron2 | HMGIY | ATF2 | ||
| NFIC | ||||||||
| EBF1 | ||||||||
| EP300 | ||||||||
| NFKB1 | ||||||||
| PAX5 | ||||||||
| chr2:203775474 | rs72936852 | 2b | WDR12 | Intron1 | AR | MAFF | ||
| chr2:203926270 | rs72934512 | 2b | NBEAL1 | Intron6 | TEAD1 | TEAD4 | ||
| TEAD3 | ||||||||
| chr8:19803092 | rs3779788 | 2b | LPL | Intron1 | TGIF | CEBPB | ||
| chr10:44757106 | rs518594 | 2b | CXCL12 | Downstream intergenic | E2A | FOXM1 | ||
| NRSE | NFIC | |||||||
| NRSF | MAX | |||||||
| EBF1 | ||||||||
| TBL1XR1 | ||||||||
| chr4:57824931 | rs7687767 | 1d | CECR6 | Sox8 | FOXA1 | |||
| chr11:5759712 | rs93138 | 1f | SWAP70 | Intron8 | ||||
| rs360136 | 1f | SWAP70 | Exon13 | |||||
| chr:457798188 | rs2227901 | 1f | REST-NOA1 | Exon6 | SPI1 | |||
| chr:1567442595 | rs17293632 | 2a | SMAD3 | Intron4 | Bach1 | SIN3A | ||
| AP-1 | TCF7L2 | |||||||
| JundM2 | TFAP2A | |||||||
| Pou1f1 | TFAP2C | |||||||
| Pou3f1 | ZNF217 | |||||||
| Sox5 | POLR2A | |||||||
| JDP2 | STAT1 | |||||||
| MXI1 | ||||||||
| TAF1 | ||||||||
| E2F1 | ||||||||
| POLR2A | ||||||||
| HDAC1 | ||||||||
| TCF7L2 | ||||||||
| MYC | ||||||||
| POLR2A | ||||||||
| ESR1 | ||||||||
| EP300 | ||||||||
| CDX2 | ||||||||
| HNF4A | ||||||||
| CEBPB | ||||||||
| EP300 | ||||||||
| FOS | ||||||||
| FOSL1 | ||||||||
| GATA3 | ||||||||
| JUNB | ||||||||
| JUN | ||||||||
| MYC | ||||||||
| NR2F2 | ||||||||
| NR3C1 | ||||||||
| RAD21 | ||||||||
| RCOR1 | ||||||||
| TCF12 | ||||||||
| JUND | ||||||||
| MAFK | ||||||||
| EGR1 | ||||||||
| MAX | ||||||||
| chr15:67441996 | rs18866316 | 2b | SMAD3 | Exon4 | AIRE | ESR1 | ||
| Elf3 | NR3C1 | |||||||
| Srf | POLR2A | |||||||
| Tcf3 | ||||||||
| Tcfap2e | ||||||||
| chr15:67448898 | rs8032739 | 2b | MTERF | Intron4 | SRF | CEBPB | ||
| FOS | ||||||||
| MYC | ||||||||
| STAT3 | ||||||||
| chr9:22098573 | rs4977574 | 2c | CDKN2BAS1 | Intron16 | AR | AR | ||
| NR3C1 | ||||||||
| NR3C2 | ||||||||
| Ar | ||||||||
| Elk-1 | ||||||||
| c-Ets-1(p54) | ||||||||
One SNP rs2075650 lies in Intron 2 of ApoEApoC1/TOMM40 with a RegulomeDB score of 1f. It is located in RREB1 DNA motif and is an eQTL for TOMM40 (Table 4).
In total 3 of 107 SMG6 associated SNPs, rs2281727, rs7217687 and rs9908888 had a score of 2b and they affect the binding of EP300. rs2281727 is a genome-wide significant SNP located in Intron 9 of SMG6. It is in binding motifs of SRY, Srf and Zfp105 and affects the binding of CREBBP, EP300, STAT3, TRIM28, MYC and RBBP5 (Table 4).
The UBE2Z region had 10 functional SNPs, including a GWAS reported SNP, UBE2Z/rs46522 (RegulomeDB score of 1f). The SNP with the most evidence of regulatory function in this locus is rs4794004 with a score of 1d. It is in DNA motif of Gata5 that alters the expression of UBE2Z and ATP5G1and affects the binding of NR3C1, IN3AK20, CREB1, TAF12, CTCF, POLR2A, USF1, FOXA1, FOXA2 and RBBP5. The other 5 SNPs in this region have a score of 1f. The remaining two regulatory SNPs, rs3744608 and rs12601672, have scores of 2a and 2b, respectively. rs3744608 is located in Intron 3 of UBE2Z and it affects the binding of large number of transcription factors (Table 4).
COL4A1-COL4A2/rs473144 is a GWAS reported SNP, achieving a RegulomeDB score of 2b. This SNP lies in Intron 3 of COL4A2 between STST3:STAT3 DNA motif and affects the binding of POLR2A and EZH2 (Table 4). ZNF259-APOA5-APOA1/rs964184 is a GWAS significant SNP with a score of 1f and is an eQTL for TAGLN. This SNP is located downstream of this gene region and is present in FOXJ2 DNA motif. Another GWAS significant SNP, ZC3HC1/rs11556924 is an exonic variant and the only functional SNP (score = 1f) in this locus; it is also an eQTL for ZC3HC1 (Table 4).
REST-NOA1/rs17087335 is in LD with two functional SNPs (rs2227901 and rs7687767 with RegulomeDB scores of 1f and 1d, respectively). rs768776 lies in DNA motif of Sox8 and affects the binding of FOXA1. SWAP70 has two functional SNPs, rs93138 and rs360136, each with a RegulomeDB score of 1f. SWAP70/rs93138 is an eQTL as evidenced in monocytes.
SMAD3 has three functional SNPs, SMAD3/rs17293632 and SMAD3/rs1866316 and MTERF1/rs8032739 with RegulomeDB scores of 2a, 2a and 2b, respectively. Both are in LD with a lead GWAS SNP (SMAD3/rs56062135). CDKN2BAS1/rs1333049 has one functional SNP(rs4977574) only with RegulomeDB score of 3c. It is a part of a gene cluster on chromosome 9p21 and it maps to Intron 16 of cyclin dependent kinase, an important regulator of cell cycle.
Discussion
Following the sequencing of human genome, a large number of SNPs have been identified that affect disease phenotypes, but their exact roles remain unclear (16). One possible explanation is that some variation affects disease expression at the transcriptional level other than at the protein level. For example, a base pair change in a transcription factor binding site may affect the binding affinity of transcription factors that consequently may alter the transcription of the related genes. These effects are indirect and may seem subtle, but their interactions with other genetic or environmental factors may result in the pathogenesis of common diseases.
Like other complex disorders, a large number of CAD associated risk variants have been discovered by multiple GWAS (12, 13, 17). ENCODE provides information regarding the functionality of human genome (18). This data requires careful interpretation and helps to define the biological function of previously termed ‘junk DNA’. Using bioinformatics tools, we may generate new hypotheses about the gene regulation of complex disorders. In this study, we have used two bioinformatics tools, SNAP and RegulomeDB, in order to identify the putative roles of CAD-associated SNPs.
We examined a total 1,200 SNPs in 54 loci implicated by GWAS, including 58 genome-wide significant SNPs. Ninety-seven SNPs were predicted to have regulatory functions with a RegulomeDB score of <3, but only 8 of them were genome-wide significant. Interestingly, all 8 genome-wide significant SNPs with suggested regulatory function are located either in intronic or intergenic regions, suggesting that these are true associations that regulate gene expression at the transcriptional level.
Among these eight GWAS reported functional SNPs, the SNP with the top RegulomeDB score was LIPA/rs2246833 (Regulome DB score = 1b). This variant is located in Intron 6 of lipase A (LIPA) and is an eQTL for the same gene which catalyzes intracellular triglyceride and hydrolyses cholesterol ester (19).
ZC3HC1/rs11556924 is a GWAS significant CAD associated SNP that returned a score of 1f. rs11556924 is a coding SNP located in the ZC3HC1 gene region encoding NIPA (Nuclear Interaction Partner of ALK) protein. This polymorphism is responsible for arginine-histidine amino acid alteration at position 363 (R363H). The SNP has been associated with essential hypertension in Finnish population (20, 21).
The CYP17A1-CNNM2-NT5C2 gene region has the highest number of regulatory SNPs, including one GWAS significant SNP, rs12413409. This locus affects diastolic blood pressure, systolic blood pressure and body mass index. All three measures are important risk factors for CAD (22). There are 25 putative regulatory SNPs in LD with rs12413409 that are located across four genes on chromosome 10 (CNNM2, NT5C2, AS3MT and BORCS7/ASMT) but affect the expression of same protein USMG5. These findings suggest that USMG5 should be investigated as an important player for CAD pathogenesis. USMG5 (upregulated during skeletal muscle growth protein 5) is also known as diabetes-associated protein in insulin-sensitive tissues that plays a crucial role in the maintenance of ATP synthase structure in mitochondria (23). Chen et al. (24) have purified this protein from bovine heart mitochondria and suggested its role in cell energy metabolism.
APOE-APOC1/TOMM40/rs2075650 is present in the TOMM40 gene region near the APOE-C1 cluster. TOMM40 encodes TOMM40 protein, which is an important subunit 40 of outer mitochondrial membrane protein complex. rs2075650 risk allele has shown an association with low levels of CRP in CAD patients (25).
rs46522, an intronic SNP in Ubiquitin-conjugating enzyme E2Z (UBE2Z) gene region returns a RegulomeDB score of 1f. This SNP is associated with CAD in Iranian and Han Chinese populations (26, 27). The exact mechanism by which genetic alteration in UBE2Z can attribute to the CAD risk is not yet clear; however, rs46522 is in strong LD with the causal SNPs in gastric inhibitory peptide (GIP) gene that encodes GIP protein, a protein that modifies the glucose and lipid metabolism potentially mediating known CAD risk factors.
ZNF259-APOA5APOA1/rs964184 is also an important regulatory SNP. ZNF259 protein polymorphism has been associated with metabolic syndrome in Chinese population. Aung et al. (28) have also shown its association with lipid levels. ZNF259 is located close to APOA5. Overexpression of APOA5 in mice reduces plasma triglyceride levels and mice lacking APOA5 have hypertriglyceridemia (29).
COL4A2/rs4773144 has been identified as functional lead SNP by RegulomeDB (score = 2b). This gene controls collagen proliferation, indicating a potential functional role in atherosclerotic plaque strengthening (30).
SMG6/rs2281727 is an intronic SNP. The potential function of SMG6 in CAD is not yet established. This gene promotes the endonuclease activity and is responsible for protection of telomere ends of chromosomes (16).
Although regulatory elements are most often found in non-coding regions of the genome, we found 5 loci with exonic regulatory SNPs (VAMP8/rs1009, CNNM2/rs943037, GIP/rs2291725, KIAA1462/rs3739998 and UBE2Z/rs15563), indicating the presence of regulatory signals inside the coding sequences as well.
USF1 is an upstream transcription factor whose binding is affected by three SNPs (GIP/rs4794004, FES/rs1894401, HHIPL1/rs28391527), suggests a potential functional link between FES, GIP and HHIPL1 (31).
RegulomeDB identified three important functional SNPs affecting CAD phenotype. Among these, REST-NOA1/rs17087335 is the lead GWAS SNP that encodes a transcription factor which suppresses the voltage gated sodium and potassium channels and it has shown to maintain vascular smooth muscle cells in non-proliferative phase (32). SWAP70/rs10840293 encodes a signaling molecule that is implicated in cell adhesion and migration and it appears to be a potential regulator of leukocyte migration and their adhesion to endothelial cells (33). SMAD3 is a major regulator of TGF-ß. A study on mice has shown that mutations in this gene lead to decreased connective tissue deposition in response to vascular injury (34).
It should be noted that 342 SNPs had returned ‘No Data’ when queried by RegulomeDB. This suggests that current evidence does not support a functional role for those variants. Our results also showed that some loci harbor markedly more regulatory SNPs as compared with other regions. We caution against interpreting this finding to meant that one region is more functionally relevant, as regions with ‘fewer’ functional SNPs may have yet to be interrogated as thoroughly and thus have fewer annotations.
Since these loci are mostly in Europeans, and only 5 of them are replicated in South Asians (35), the findings may not be as relevant to other populations as they are to Europeans as genetic effects can differ across populations. The cause of this varying association with disease phenotype may be the ethnic admixture resulting in population stratification. It is also noteworthy that robust associations of variants with different diseases have been reported in Europeans while other populations (Africans, Asians and Hispanics) failed to demonstrate those associations (36, 37).
Though the cellular mechanisms underlying CAD pathogenesis are established, the molecular basis is not yet agreed upon. Comprehending the molecular basis of disease is crucial before pathogenesis is completely described. The study has identified 97 regulatory SNPs associated with CAD. In summary, our results highlight the importance of considering both disease-associated SNPs and those SNPs in LD, as well as the regulatory function of these SNPs to help identify the causal genetic mechanisms of CAD. The methods which we have implemented here can inform planning of more complete and better directed functional genomic studies.
Supplementary data
Supplementary data are available at Database Online.
Funding
This study was partially supported by Higher Education Commission of Pakistan and the US National Institutes of Health grants (AG030653 and AG041718).
Conflict of interest. None declared.
References
Author notes
Citation details: Cheema,A.N., Rosenthal,S.L., and Kamboh,M.I. Proficiency of data interpretation: Identification of signaling SNPs/specific loci for coronary artery disease. Database (2017) Vol. 2017: article ID bax078; doi:10.1093/database/bax078



