-
PDF
- Split View
-
Views
-
Cite
Cite
Dina Vishnyakova, Emilie Pasche, Patrick Ruch, Using binary classification to prioritize and curate articles for the Comparative Toxicogenomics Database, Database, Volume 2012, 2012, bas050, https://doi.org/10.1093/database/bas050
Close - Share Icon Share
Abstract
We report on the original integration of an automatic text categorization pipeline, so-called ToxiCat (Toxicogenomic Categorizer), that we developed to perform biomedical documents classification and prioritization in order to speed up the curation of the Comparative Toxicogenomics Database (CTD). The task can be basically described as a binary classification task, where a scoring function is used to rank a selected set of articles. Then components of a question-answering system are used to extract CTD-specific annotations from the ranked list of articles. The ranking function is generated using a Support Vector Machine, which combines three main modules: an information retrieval engine for MEDLINE (EAGLi), a gene normalization service (NormaGene) developed for a previous BioCreative campaign and finally, a set of answering components and entity recognizer for diseases and chemicals. The main components of the pipeline are publicly available both as web application and web services. The specific integration performed for the BioCreative competition is available via a web user interface at http://pingu.unige.ch:8080/Toxicat.
Introduction
We report on the original integration of an automatic text categorization pipeline, so-called ToxiCat (Toxicogenomic Categorizer), that we developed to perform biomedical documents classification and prioritization in order to speed up the curation of the Comparative Toxicogenomics Database (CTD). The task can be basically described as a binary classification task, where a scoring function is used to rank a selected set of articles. Then components of a question-answering system are used to extract CTD-specific annotations from the ranked list of articles. The ranking function is generated using a Support Vector Machine (SVM), which combines three main modules: an information retrieval engine for MEDLINE (EAGLi), a gene normalization (GN) service (NormaGene) developed for a previous BioCreative campaign and finally, a set of answering components and entity recognizer for diseases and chemicals. The main components of the pipeline are publicly available both as web application and web services. The specific integration performed for the BioCreative competition is available via a web user interface at http://pingu.unige.ch:8080/Toxicat.
Biocuration pipeline
Biocuration is a complex task, which requires domain expertise and specific training. The task, when performed by a professional curator or by an automatic annotation system, can be simplified into the following workflow (R.S.R.E.N.):
Retrieval of documents given a particular query (e.g. proteins, chemicals) in a particular document repository (MEDLINE, patent library, PubMed Central, etc.).
Selection of articles: a subset of articles is chosen by the curator, usually based on the title and the abstract.
Reading of a particular article (or passage retrieval for an automat): the full-text article is read by the biologist.
Extraction of information: a particular passage is analyzed to obtain a representation of the level of entities such as proteins, diseases, methods used to generate a particular result (e.g. yeast two-hybrid) and evidence codes (e.g. automatic inference, direct interactions).
Normalization of the extracted information: the biologist transforms a particular passage into a normalized identifier (e.g. a disease for CTD) or a set of normalized descriptors (e.g. several protein identifiers and an interaction type for protein–protein interaction databases such as IntAct).
Feed-back: this step is optional; it aims at using the generated annotation to improve or refine the search initiated in Step 1.
It is worth observing that Step 1 is directly dependent on the user interface of the curation platform. Various user interaction models can be designed here: interactive search (e.g. PubMed, EBIMed, EAGLi, etc.) or batch search, where the curator receives regular (daily, weekly, etc.) alerts and notifications. In the alerting model, queries must have been previously registered by the curator. Moreover, in many biocuration systems, Steps 1 and 2, which are sometimes called ‘triage’ tasks, are performed by professional biologists. Thus, functional annotation systems, like those originally designed during BioCreative I (1), help curators to assign Gene Ontology descriptors to gene products; only Steps 3–5 are performed by a computer, see e.g. the GO categorizer (2). For the BioCreative 2012 evaluation campaign, the organizers provided a flat list of PMIDs so most systems did not need to provide any retrieval functionalities (Step 1). Step 6 is also often ignored by designers of text mining systems for biocuration and was not mandatory for the competition.
The system we designed tentatively covered all steps. A graphic user interface (GUI) has been designed for the sake of the BioCreative competition; however, such a GUI must be regarded as a basic demonstrator; see Table 1 for an overview of the basic integration we designed. This integration step is obviously critical for the success of a curation system. However, it goes far beyond the scope of our report, which focuses on the integration and evaluation of a set of text mining services. The light integration we prepared for BioCreative is thus based on the existing EAGLi platform, which is used to acquire PMIDs and to further answer questions resulting from an automatic annotation process. Table 1 explains how the curation workflow has been instantiated in our CTD curation system (ToxiCat). The construction and evaluation of the ToxiCat binary classifier is then the main subject of this report since other components have been described elsewhere: EAGLi search (3), EAGLi’s question-answering (3), EAGLi’s Keyword extractor (3), GOCat (2) and NormaGene (4).
Components used to generate the ToxiCat system that was designed during BioCreative 2012 to curate the Comparative Toxicogenomics Database
| . | Assisted EAGLi or PubMed . | Assisted ToxiCat’s binary classifier . | Keyword in context (EAGLi) . | Named-Entity recognition ad hoc NormaGeneGOCat . | Categorization EAGL, NormaGene . | EAGLi Question- Answering . |
|---|---|---|---|---|---|---|
| Retrieval | X | |||||
| Selection | X | |||||
| Reading | X | |||||
| Extraction | X | |||||
| Normalization | X | |||||
| Feed-back | X | X |
| . | Assisted EAGLi or PubMed . | Assisted ToxiCat’s binary classifier . | Keyword in context (EAGLi) . | Named-Entity recognition ad hoc NormaGeneGOCat . | Categorization EAGL, NormaGene . | EAGLi Question- Answering . |
|---|---|---|---|---|---|---|
| Retrieval | X | |||||
| Selection | X | |||||
| Reading | X | |||||
| Extraction | X | |||||
| Normalization | X | |||||
| Feed-back | X | X |
Components used to generate the ToxiCat system that was designed during BioCreative 2012 to curate the Comparative Toxicogenomics Database
| . | Assisted EAGLi or PubMed . | Assisted ToxiCat’s binary classifier . | Keyword in context (EAGLi) . | Named-Entity recognition ad hoc NormaGeneGOCat . | Categorization EAGL, NormaGene . | EAGLi Question- Answering . |
|---|---|---|---|---|---|---|
| Retrieval | X | |||||
| Selection | X | |||||
| Reading | X | |||||
| Extraction | X | |||||
| Normalization | X | |||||
| Feed-back | X | X |
| . | Assisted EAGLi or PubMed . | Assisted ToxiCat’s binary classifier . | Keyword in context (EAGLi) . | Named-Entity recognition ad hoc NormaGeneGOCat . | Categorization EAGL, NormaGene . | EAGLi Question- Answering . |
|---|---|---|---|---|---|---|
| Retrieval | X | |||||
| Selection | X | |||||
| Reading | X | |||||
| Extraction | X | |||||
| Normalization | X | |||||
| Feed-back | X | X |
Data and methods
Data overview
BioCreative 2012 proposes to explore how text mining methods can successfully be applied to practically help biocuration of a large molecular biology knowledge base. The main objective of the Triage-I task is to explore how a set of MEDLINE records, directly retrieved from PubMed using the name of a particular chemical compound, can be ranked to prioritize the most relevant articles. In addition to the prioritized list of PMIDs, competitors are also asked to provide additional annotations of interest to maintain the CTD with the interacting entities (small molecules and gene products) and the pathologies likely to reflect the toxicity of the chemical compound; see (5) for more detailed information about all tasks of BioCreative 2012 and CTD.
The organizers provided a set of 1725 abstracts for training. This set was triaged and curated, but it is worth observing that CTD curators also use full-text articles to annotate the database. Approximately more than half of these articles contain no information about the chemical compound and/or the genes they are supposed to annotate. Symmetrically, less than half of the articles contain information about both a gene and a chemical, and only about an eighth of the articles contain information about diseases. The distribution of entity types in the benchmark is shown in Table 2.
| Entity name . | Number of articles . |
|---|---|
| Chemical compounds | 735 |
| Genes | 583 |
| Diseases | 204 |
| Co-occurrence of genes and chemical compounds | 542 |
| Input chemical compound in titles | 381 |
| Entity name . | Number of articles . |
|---|---|
| Chemical compounds | 735 |
| Genes | 583 |
| Diseases | 204 |
| Co-occurrence of genes and chemical compounds | 542 |
| Input chemical compound in titles | 381 |
| Entity name . | Number of articles . |
|---|---|
| Chemical compounds | 735 |
| Genes | 583 |
| Diseases | 204 |
| Co-occurrence of genes and chemical compounds | 542 |
| Input chemical compound in titles | 381 |
| Entity name . | Number of articles . |
|---|---|
| Chemical compounds | 735 |
| Genes | 583 |
| Diseases | 204 |
| Co-occurrence of genes and chemical compounds | 542 |
| Input chemical compound in titles | 381 |
The data were distributed in a set of files that included eight curated CTD entries describing the following chemical compounds: raloxifene, aniline, amasacrine, doxorubicin, aspartame, quercetin, 2-acetylaminofluorene and indomethacin (5). Our preliminary experiments show that using only the four chemicals shown in Table 3 performed better than using the eight compounds. This group of four chemical compounds is annotated with 1059 articles. The distribution of positive and negative instances in this subset is nearly balanced (see Table 3).
| Chemical compound name . | Number of articles per chemical compound . | %Positive articles in the sample . |
|---|---|---|
| Raloxifene | 270 | 60 |
| 2-Acetylaminofluorene | 178 | 45.5 |
| Amsacrine | 69 | 53 |
| Quercetimin | 542 | 77 |
| Chemical compound name . | Number of articles per chemical compound . | %Positive articles in the sample . |
|---|---|---|
| Raloxifene | 270 | 60 |
| 2-Acetylaminofluorene | 178 | 45.5 |
| Amsacrine | 69 | 53 |
| Quercetimin | 542 | 77 |
| Chemical compound name . | Number of articles per chemical compound . | %Positive articles in the sample . |
|---|---|---|
| Raloxifene | 270 | 60 |
| 2-Acetylaminofluorene | 178 | 45.5 |
| Amsacrine | 69 | 53 |
| Quercetimin | 542 | 77 |
| Chemical compound name . | Number of articles per chemical compound . | %Positive articles in the sample . |
|---|---|---|
| Raloxifene | 270 | 60 |
| 2-Acetylaminofluorene | 178 | 45.5 |
| Amsacrine | 69 | 53 |
| Quercetimin | 542 | 77 |
Methods
We designed a SVM classifier for the binary classification of articles with two classes: relevant for curation and not relevant for curation. All our experiments and developments use the libSVM package (6). The classifier returns a Boolean value together with a class estimate, which directly expresses the probability to belong either to the positive or the negative class. The features, which were selected to build the classification model, are shown in Table 4. An F-score is provided, which expresses the respective contribution of each feature to the binary classification model, as described in (8).
| Features . | FScore . | Source . |
|---|---|---|
| Normalized input chemical compound in MeSH terms | 0.008 | EAGLi |
| Journal name relevant for CTD task | 0.1593 | |
| Appearance of ‘pharmacology’, ‘toxicity’, ‘drug therapy’, ‘metabolism’, ‘drug effects’, ‘chemistry’ and ‘chemical synthesis’ as the main MeSH terms of the article | 0.0672 | |
| Input chemical compound in the abstract | 0.025 | Ad hoc keywords Recognizer |
| Input chemical compound in the title | 0.028 | |
| Chemical compounds in an abstract | 0.023 | |
| Frequency of chemical compounds in an abstract | 0.0009 | |
| Frequency of input chemical in an abstract | 0.0036 | |
| Input chemical compound detected in first three sentencesa | 0.0027 | |
| Diseases in the abstract | 0.0111 | |
| Chemical compounds and genes in an abstract | 0.072 | NormaGene |
| Co-occurrence of genes and chemical compounds in a sentence | 0.0036 | |
| Co-occurrence of main chemical and genes in a sentence | 0.0001 | |
| Sum of score of every feature | 0.04 | EAGLi + ad hoc keywords Recognizer + NormaGene |
| Features . | FScore . | Source . |
|---|---|---|
| Normalized input chemical compound in MeSH terms | 0.008 | EAGLi |
| Journal name relevant for CTD task | 0.1593 | |
| Appearance of ‘pharmacology’, ‘toxicity’, ‘drug therapy’, ‘metabolism’, ‘drug effects’, ‘chemistry’ and ‘chemical synthesis’ as the main MeSH terms of the article | 0.0672 | |
| Input chemical compound in the abstract | 0.025 | Ad hoc keywords Recognizer |
| Input chemical compound in the title | 0.028 | |
| Chemical compounds in an abstract | 0.023 | |
| Frequency of chemical compounds in an abstract | 0.0009 | |
| Frequency of input chemical in an abstract | 0.0036 | |
| Input chemical compound detected in first three sentencesa | 0.0027 | |
| Diseases in the abstract | 0.0111 | |
| Chemical compounds and genes in an abstract | 0.072 | NormaGene |
| Co-occurrence of genes and chemical compounds in a sentence | 0.0036 | |
| Co-occurrence of main chemical and genes in a sentence | 0.0001 | |
| Sum of score of every feature | 0.04 | EAGLi + ad hoc keywords Recognizer + NormaGene |
The features are grouped according to the sources that produced them.
aWe tried to use features generated by an argumentative classifier (7), but preliminary experiments were inconclusive.
| Features . | FScore . | Source . |
|---|---|---|
| Normalized input chemical compound in MeSH terms | 0.008 | EAGLi |
| Journal name relevant for CTD task | 0.1593 | |
| Appearance of ‘pharmacology’, ‘toxicity’, ‘drug therapy’, ‘metabolism’, ‘drug effects’, ‘chemistry’ and ‘chemical synthesis’ as the main MeSH terms of the article | 0.0672 | |
| Input chemical compound in the abstract | 0.025 | Ad hoc keywords Recognizer |
| Input chemical compound in the title | 0.028 | |
| Chemical compounds in an abstract | 0.023 | |
| Frequency of chemical compounds in an abstract | 0.0009 | |
| Frequency of input chemical in an abstract | 0.0036 | |
| Input chemical compound detected in first three sentencesa | 0.0027 | |
| Diseases in the abstract | 0.0111 | |
| Chemical compounds and genes in an abstract | 0.072 | NormaGene |
| Co-occurrence of genes and chemical compounds in a sentence | 0.0036 | |
| Co-occurrence of main chemical and genes in a sentence | 0.0001 | |
| Sum of score of every feature | 0.04 | EAGLi + ad hoc keywords Recognizer + NormaGene |
| Features . | FScore . | Source . |
|---|---|---|
| Normalized input chemical compound in MeSH terms | 0.008 | EAGLi |
| Journal name relevant for CTD task | 0.1593 | |
| Appearance of ‘pharmacology’, ‘toxicity’, ‘drug therapy’, ‘metabolism’, ‘drug effects’, ‘chemistry’ and ‘chemical synthesis’ as the main MeSH terms of the article | 0.0672 | |
| Input chemical compound in the abstract | 0.025 | Ad hoc keywords Recognizer |
| Input chemical compound in the title | 0.028 | |
| Chemical compounds in an abstract | 0.023 | |
| Frequency of chemical compounds in an abstract | 0.0009 | |
| Frequency of input chemical in an abstract | 0.0036 | |
| Input chemical compound detected in first three sentencesa | 0.0027 | |
| Diseases in the abstract | 0.0111 | |
| Chemical compounds and genes in an abstract | 0.072 | NormaGene |
| Co-occurrence of genes and chemical compounds in a sentence | 0.0036 | |
| Co-occurrence of main chemical and genes in a sentence | 0.0001 | |
| Sum of score of every feature | 0.04 | EAGLi + ad hoc keywords Recognizer + NormaGene |
The features are grouped according to the sources that produced them.
aWe tried to use features generated by an argumentative classifier (7), but preliminary experiments were inconclusive.
Features can be split within three subsets. The first feature set contains information about MeSH terms of articles extracted from the MEDLINE library, which is locally stored and indexed by the EAGLi’s engine. The use of EAGLi has two main advantages when compared with PubMed’s e-Utilities: (i) the response time is significantly improved from about 1 s per PMID to an average of 50 ms for EAGLi, which results in an overall processing time at least one order of magnitude faster; (ii) recently published articles are not yet indexed with MeSH, while EAGLi offers the possibility to automatically index those articles with a modest processing time (∼200 ms); see (2) for a presentation of the MeSH assignment system and (9) for a comparison against similar systems such as MetaMap. According to our observations, curated articles are usually indexed with major headings such as pharmacology, toxicity, drug therapy, metabolism, drug effects, chemistry and chemical synthesis. Very often, the indexing with MeSH also normalizes the name of the main chemical with a unique identifier (or preferred term) discussed in the article (i.e. raloxifene or amsacrine). The second subset of features is obtained via the NormaGene named-entity normalizer (4, 10). This gene and protein named-entity recognizer was developed for the BioCreative III task to address the GN task (4). Like other named-entity recognizer, it identifies the boundaries of the gene and protein name, but it also attempts to assign a unique identifier at the level of a UniProt or Entrez-Gene sequence. Thus, NormaGene also attempts to recognize, when possible, what organisms are mentioned in the article to ultimately link a gene/protein name with a unique sequence. When the species are not explicitly mentioned, NormaGene attempts to derive it from other textual entities such as cell lines or gene products. Internally, NormaGene is able to recognize all gene candidates stored in the Gene and Protein Synonyms DataBase (GPSDB) (11), as well as all species stored in NEWT (12), which is appropriate to annotate contents for UniProt/SwissProtKB but which does exceed the coverage of CTD. The internal gene and gene product dictionaries of NormaGene are therefore reduced to curate CTD. Finally, results returned by NormaGene are compared with the CTD genes controlled vocabulary to further reduce the list of results. The controlled vocabulary of CTD contains over 257 000 NCBI genes’ identifiers and over 479 000 genes’ names including synonyms. If the entities recognized by NormaGene are found in the CTD genes’ vocabulary, then we extract all synonyms based on the approved genes ID and match them against the abstract. Indeed, gene and protein identifiers suggested by NormaGene cannot always be explicitly found in the body of the input document as NormaGene uses a generative model, which exploits also functional similarities (13) and not only textual similarities. Gene names used by CTD are imported from EntrezGene but unlike Entrez-Gene, a gene product in CTD is mostly concerned with human-related toxicology, which simplifies the gene recognition process.
The third set of features is an ad hoc keyword recognizer for diseases and chemicals. This keyword recognizer is based on the controlled vocabularies provided by CTD. We discovered that CTD vocabularies for chemicals and diseases contain several descriptors, which seems irrelevant for the curation task. However, the description of CTD vocabularies (see the ‘CTD curation overview’ on http://www.biocreative.org/tasks/bc-workshop-2012/triage/) explains that several branches of the original MeSH vocabulary were pruned from CTD’s chemical and disease vocabularies (14) because of their weak relevance for CTD. Nevertheless, we discovered that in the vocabularies provided by the organizers, we have all these branches. It was therefore challenging to decide a priori which descriptors should have been excluded or not. For this task, we decided to rely on the UMLS Metathesaurus. For both chemical and disease entities, we created a Word-Sense Disambiguator (WSD) based on the UMLS Semantic Types (15). We remove non-relevant types of chemicals and diseases as listed in Table 5. Further, in order to eliminate common English words from the list of candidates, we created a common English word recognizer based on a general-purpose English corpora. Unspecific disease and chemical names were thus discarded.
| UMLS Semantic Type . | Name . |
|---|---|
| T023 | Body part, organ or organ component |
| T031 | Body substance |
| T037 | Injury or poisoning |
| T046 | Pathologic function |
| T073 | Manufactured object |
| T080 | Qualitative concept |
| T081 | Quantitative concept |
| T086 | Nucleotide sequence |
| T087 | Amino acid sequence |
| T088 | Carbohydrate sequence |
| T103 | Compounds or substances of definite molecular composition. |
| T114 | Nucleic acid, nucleoside or nucleotide |
| T116 | Amino acid, peptide or protein |
| T118 | Carbohydrate |
| T119 | Lipid |
| T120 | Chemical viewed functionally |
| T123 | Biologically active substance |
| T125 | Hormone |
| T126 | Enzyme |
| T127 | Vitamin |
| T129 | Immunologic factor |
| T168 | Food |
| T196 | Element, ion or isotope |
| T197 | Inorganic chemical |
| UMLS Semantic Type . | Name . |
|---|---|
| T023 | Body part, organ or organ component |
| T031 | Body substance |
| T037 | Injury or poisoning |
| T046 | Pathologic function |
| T073 | Manufactured object |
| T080 | Qualitative concept |
| T081 | Quantitative concept |
| T086 | Nucleotide sequence |
| T087 | Amino acid sequence |
| T088 | Carbohydrate sequence |
| T103 | Compounds or substances of definite molecular composition. |
| T114 | Nucleic acid, nucleoside or nucleotide |
| T116 | Amino acid, peptide or protein |
| T118 | Carbohydrate |
| T119 | Lipid |
| T120 | Chemical viewed functionally |
| T123 | Biologically active substance |
| T125 | Hormone |
| T126 | Enzyme |
| T127 | Vitamin |
| T129 | Immunologic factor |
| T168 | Food |
| T196 | Element, ion or isotope |
| T197 | Inorganic chemical |
| UMLS Semantic Type . | Name . |
|---|---|
| T023 | Body part, organ or organ component |
| T031 | Body substance |
| T037 | Injury or poisoning |
| T046 | Pathologic function |
| T073 | Manufactured object |
| T080 | Qualitative concept |
| T081 | Quantitative concept |
| T086 | Nucleotide sequence |
| T087 | Amino acid sequence |
| T088 | Carbohydrate sequence |
| T103 | Compounds or substances of definite molecular composition. |
| T114 | Nucleic acid, nucleoside or nucleotide |
| T116 | Amino acid, peptide or protein |
| T118 | Carbohydrate |
| T119 | Lipid |
| T120 | Chemical viewed functionally |
| T123 | Biologically active substance |
| T125 | Hormone |
| T126 | Enzyme |
| T127 | Vitamin |
| T129 | Immunologic factor |
| T168 | Food |
| T196 | Element, ion or isotope |
| T197 | Inorganic chemical |
| UMLS Semantic Type . | Name . |
|---|---|
| T023 | Body part, organ or organ component |
| T031 | Body substance |
| T037 | Injury or poisoning |
| T046 | Pathologic function |
| T073 | Manufactured object |
| T080 | Qualitative concept |
| T081 | Quantitative concept |
| T086 | Nucleotide sequence |
| T087 | Amino acid sequence |
| T088 | Carbohydrate sequence |
| T103 | Compounds or substances of definite molecular composition. |
| T114 | Nucleic acid, nucleoside or nucleotide |
| T116 | Amino acid, peptide or protein |
| T118 | Carbohydrate |
| T119 | Lipid |
| T120 | Chemical viewed functionally |
| T123 | Biologically active substance |
| T125 | Hormone |
| T126 | Enzyme |
| T127 | Vitamin |
| T129 | Immunologic factor |
| T168 | Food |
| T196 | Element, ion or isotope |
| T197 | Inorganic chemical |
The general architecture of the ToxiCat workflow is shown in Figure 1. Articles data such as the PMID, the abstract and the journal name are passed to ToxiCat. The PMID is used to query EAGLi’s services in order to retrieve all MeSH terms of the article. The abstract of an article is also passed to the ad hoc keyword recognizer to detect chemicals and diseases candidates. Those candidates are then filtered by the common English word filter and finally by the WSD. In parallel, NormaGene detects the genes’ names in the abstract and pass them to the CTD genes Control Vocabulary, which is going to filter out not relevant genes. The remaining gene identifiers are sent to the ad hoc synonyms recognizer, to detect all synonym names in the abstract. The journal Mapper checks the name of the journal against a list of domain-relevant journal names. Finally, the resulting bag of features is processed by the SVM classifier, which returns a score. This score directly expresses the probability that the article is relevant or not to be further annotated for CTD. The parameters of the SVM classification model are obtained using 10-fold cross-validation. We tested the model with a polynomial kernel and a RBF kernel, but the results were not significantly better than with the linear kernel we finally selected.
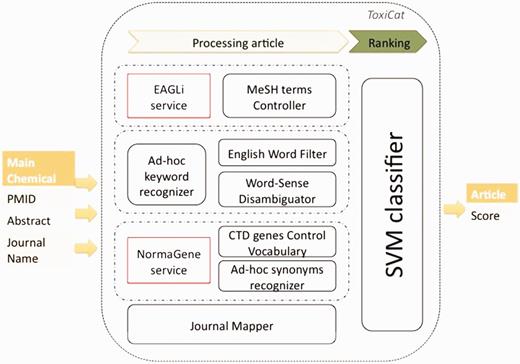
Workflow of ToxiCat and dependencies with existing online services.
In Figures 2–5, we show an example of the full biocuration process with the ToxiCat web interface.
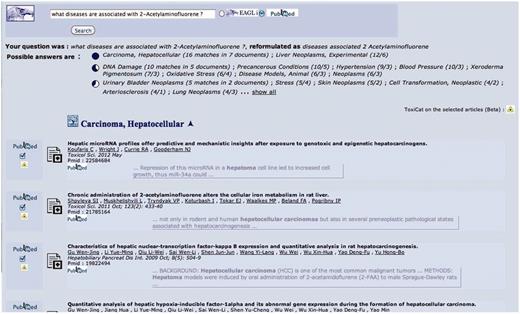
This is the starting point and also—if the user decides to click on the final questions generated by the system, see Figure 5—the end point of the search and annotation process. Here, the user can select some PMIDs, which will be then sent to ToxiCat (Figure 3) to be prioritized (Figure 4) and finally processed to generate an annotation (Figures 4 and 5).

In this figure, 2-acetylaminofluorene is provided as input chemical compound together with a list of articles (PMIDs) selected in Figure 2. Users can go directly to this page if the PMIDs have been obtained from other sources.
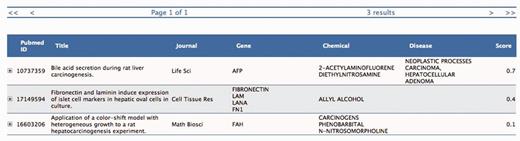
The three selected PMIDs are ranked according to the statistical estimate (Score) computed by the SVM binary classifier. Each information extraction module (Gene, Chemical, Disease) provides here a list of descriptors for each PMID together with some meta-data (Journal name, Title, etc.), which are used as features by the classifier.
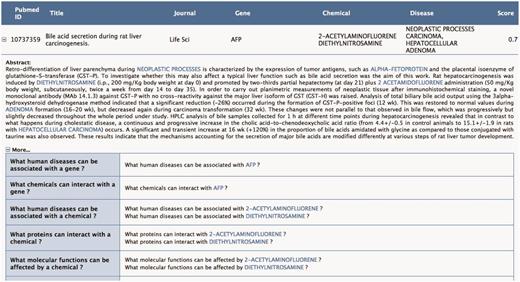
The user can request to visualize in the abstract the context of the annotation proposed in Figure 4. Toxicat tags genes/proteins, chemicals and diseases in the abstract, providing a direct link to the CTD database for each of these entities. Finally, ToxiCat generates a set of questions (‘More…’) based on the entities that were earlier extracted. Optionally, the user can then return to the EAGLi’s question-answering engine to obtain more information. The user can also obtain a list of Gene Ontology descriptors proposed by the GOCat Gene Ontology categorizer (http://eagl.unige.ch/GOCat/) based on the content of the PMID, cf. last line of the table.
Results and conclusion
The results of ToxiCat (Group 120), computed on the official data provided by BioCreative 2012’s organizers using official metrics, are shown in Table 6. This table provides two types of results:
Triage results: the mean average precision obtained when ranking documents (or MAP score).
Entity recognition results: the recalls obtained when attempting to recognize disease entities (Curated Disease Hit Rate), chemical entities (Curated Chemical Hit Rate) and gene products (Curated Gene Hit Rate).
In Table 6, ToxiCat shows competitive results in the following subtasks: relevance ranking, disease curation and chemical entity curation. From the results in Figure 6, it is also possible to see that recognition of genes and diseases in articles is usually more difficult than recognition of chemical entities. According to the results in Table 6, our gene recognition method (NormaGene) scores relatively low compared with the ad hoc chemical and disease recognition methods we developed for the competition. This result suggests that it is difficult to accurately customize a general purpose gene normalizer for a specific database curation task, although NormaGene obtained competitive results on the cross-species BioCreative III’s GN task (4).
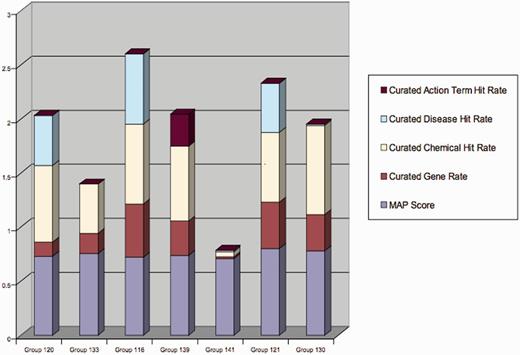
Aggregated scores of all participants in Track-I. ToxiCat is denoted under Team 120 (5).
Comparative results of ToxiCat (Team 120) for the Task-I of BioCreative 2012
| Chemical/number of articles . | Intermediate MAP score . | Curated Gene Hit Rate . | Curated Chemical Hit Rate . | Curated Disease Hit Rate . |
|---|---|---|---|---|
| Urethane/204 | 0.637 | 0.08 | 0.705 | 0.3 |
| Phenacetin/86 | 0.831 | 0.203 | 0.676 | 0.5 |
| Cyclophosphamide/154 | 0.716 | 0.117 | 0.747 | 0.582 |
| Chemical/number of articles . | Intermediate MAP score . | Curated Gene Hit Rate . | Curated Chemical Hit Rate . | Curated Disease Hit Rate . |
|---|---|---|---|---|
| Urethane/204 | 0.637 | 0.08 | 0.705 | 0.3 |
| Phenacetin/86 | 0.831 | 0.203 | 0.676 | 0.5 |
| Cyclophosphamide/154 | 0.716 | 0.117 | 0.747 | 0.582 |
Comparative results of ToxiCat (Team 120) for the Task-I of BioCreative 2012
| Chemical/number of articles . | Intermediate MAP score . | Curated Gene Hit Rate . | Curated Chemical Hit Rate . | Curated Disease Hit Rate . |
|---|---|---|---|---|
| Urethane/204 | 0.637 | 0.08 | 0.705 | 0.3 |
| Phenacetin/86 | 0.831 | 0.203 | 0.676 | 0.5 |
| Cyclophosphamide/154 | 0.716 | 0.117 | 0.747 | 0.582 |
| Chemical/number of articles . | Intermediate MAP score . | Curated Gene Hit Rate . | Curated Chemical Hit Rate . | Curated Disease Hit Rate . |
|---|---|---|---|---|
| Urethane/204 | 0.637 | 0.08 | 0.705 | 0.3 |
| Phenacetin/86 | 0.831 | 0.203 | 0.676 | 0.5 |
| Cyclophosphamide/154 | 0.716 | 0.117 | 0.747 | 0.582 |
We evaluated the effectiveness of our ad hoc terms recognizer for diseases using usual recall and precision metrics. Our methods achieved a precision of 95% and a recall of 92% when tagging diseases in the training sample. On the training data, our optimal model obtained an accuracy of 80.5%. Then, we applied this model on the official data and obtained an accuracy of 77%, which suggests some moderate overfitting phenomena of our disease recognizer.
Figure 6 shows the results of the competition for each participating team. Our team (Team 120) was ranked #3 when adding together entity recall and mean average precision. Such a result seems satisfying considering that our main work was to integrate existing components. However, it is worth to observe that the official evaluation was mainly driven by recall; therefore, it was theoretically possible to achieve top performances by providing low-quality precision. Thus, Team 116 apparently reports on competitive results, while in fact it underperforms most other competitors when looking at precision (mean average precision). This observation suggests that results submitted by this team contained more relevant descriptors but it is also possible that the results obtained by this team do contain more irrelevant descriptors!
Although current results seem suggesting that text mining can effectively help curators’ tasks by providing access to more relevant contents, it is worth noticing that the effectiveness of ToxiCat is obtained by specializing some of the components. Indirectly, we defined an ‘average user’, while the real curation work might request a more complex design. When designing the system, we somehow customize a rather generic text processing pipeline (a search engine, EAGLi, a gene named-entity normalizer, NormaGene and several terminological resources such as GPSDB) to answer the specific needs of CTD. Such a step seems both rationale and empirically effective; however, it questions the role of the end-user platform. Indeed, if the system must help the professional annotator to curate CTD by basically speeding up prioritization of articles, then a system like ToxiCat might be suitable. On the opposite, if the system should help curating non-usual contents or novel chemical products, then the system is very likely inappropriate. Ultimately, if the system was to be used as the sole capturing tool for CTD curators, then it may hinder the annotation of new interacting genes, which are not yet listed in CTD as by design non-CTD genes are penalized by the system.
In conclusion, ToxiCat showed competitive performance, in particular for the recognition of disease and chemical compounds, but such an observation must be handled carefully since precision of the annotation has not been officially evaluated. More informative, the mean average precision (MAP), which measures the ranking effectiveness of ToxiCat, is also fairly competitive. MAP score showed that the selected SVM model produced promising results. Interestingly, the identification of pathologies seems nearly as difficult as the recognition of genes and gene products, while compared with gene and protein names such entities have been largely neglected. Finally, we plan to further investigate how a question-answering engine can be integrated into a biocuration pipeline, in particular to address situations where training data are not available.
Funding
The work presented in this article has been partially supported by the DebugIT project (http://www.debugit.eu/). DebugIT is funded by the European Community’s Seventh Framework Programme under grant agreement n° FP7–217139, which is gratefully acknowledged.
Conflict of interest. None declared.



