-
PDF
- Split View
-
Views
-
Cite
Cite
Jose Cruz-Toledo, Maureen McKeague, Xueru Zhang, Amanda Giamberardino, Erin McConnell, Tariq Francis, Maria C. DeRosa, Michel Dumontier, Aptamer base: a collaborative knowledge base to describe aptamers and SELEX experiments, Database, Volume 2012, 2012, bas006, https://doi.org/10.1093/database/bas006
Close - Share Icon Share
Abstract
Over the past several decades, rapid developments in both molecular and information technology have collectively increased our ability to understand molecular recognition. One emerging area of interest in molecular recognition research includes the isolation of aptamers. Aptamers are single-stranded nucleic acid or amino acid polymers that recognize and bind to targets with high affinity and selectivity. While research has focused on collecting aptamers and their interactions, most of the information regarding experimental methods remains in the unstructured and textual format of peer reviewed publications. To address this, we present the Aptamer Base, a database that provides detailed, structured information about the experimental conditions under which aptamers were selected and their binding affinity quantified. The open collaborative nature of the Aptamer Base provides the community with a unique resource that can be updated and curated in a decentralized manner, thereby accommodating the ever evolving field of aptamer research.
Database URL:http://aptamer.freebase.com
Introduction
Over the past several decades, rapid developments in both molecular and information technology have collectively increased our ability to understand molecular recognition (1), leading to major implications for drug discovery (2,3). For example, in vitro screening techniques and adaptive molecular evolution methods, such as phage display (4), have made it easier to rapidly screen enormous molecular libraries to find promising binding ligands (5). Concurrently, advances in biomedical informatics have made it possible to harness the power of large datasets and make influential predictions for molecular recognition (6). Together, the scalability of information systems coupled with the massive reduction in the cost, efficiency and time of techniques such as DNA sequencing (7) will continue to support the exponential growth of information in the field of molecular biology (8–10).
One emerging area of interest in the quest to understand molecular recognition includes the isolation of aptamers. Aptamers are single-stranded nucleic acid or amino acid polymers that recognize and bind to targets with high affinity and selectivity. Nucleic acid aptamers are typically isolated from large combinatorial libraries containing approximately 1015 different sequences. This in vitro selection procedure is usually performed using the Systematic Evolution of Ligands by Exponential enrichment (SELEX) process (11,12). The resulting selected aptamers are at times able to bind to their cognate ligands with dissociation constants in the picomolar range (13), and can be selected for a wide variety of targets including small molecules (14), organic dyes (15), toxins (16), proteins (17,18) and whole cells (19). As a result, aptamers have emerged as attractive molecular recognition agents that rival antibodies in therapeutic (20,21), diagnostic (22) and sensing (23) applications.
While several efforts have focused on collecting aptamers and their interactions (24–26), most of the information regarding experimental methods still remains in the unstructured and textual format of peer reviewed publications. Furthermore, access to the databases is normally limited to HTML forms, thereby hindering the type and number of queries that can be posed against these datasets. Additionally, major sequence data providers such as GenBank (27), EMBL (28) and DDBJ (29) do not maintain a list of artificially created sequences. Consequently, the community lacks a data resource in which information about aptamers, their sequences and the experimental conditions used in their selection can be stored and queried.
To address this, we present the Aptamer Base, a database that provides detailed, structured information about the experimental conditions under which aptamers were selected and their binding affinity quantified. The open collaborative nature of the Aptamer Base provides the community with a unique resource that can be updated and curated in a decentralized manner, thereby accommodating the ever evolving field of aptamer research.
Materials and Methods
The Aptamer Base was created using Freebase (http://www.freebase.com), a free, openly licensed community-built resource for structured data that provides information for more than 22 million topics. Freebase organizes over 360 million facts into bases, which are collections of thematically related topics. The Aptamer Base is one such collection that has been built by a group of expert curators who manually extracted information on over 157 SELEX-based experiments from the primary literature published between 1990 and 2006. Entries in the literature from 2006 to present are added on a weekly basis.
The Aptamer Base data model
Based on the entity-relationship model (30), data in Freebase is stored as a structured graph in which ‘topics’ represent types or individuals. Every topic is identified by a unique identifier and is accessible by web browser through a Uniform Resource Locator (URL). Topics that represent types can be organized into a type hierarchy using the IS A relationship. For instance, the RNA Aptamer is a kind of Aptamer and a kind of RNA, and has access to properties from both. A type can be related to a property with the HAS A relationship, and the values of the properties can be restricted to a particular type. For example, the Dissociation Constant type HAS A property called ‘is dissociation constant of’ that links to topics only of the Interactor type, as in http://www.freebase.com/view/m/0cjchc8.
The Aptamer Base data model (Figure 1) is focused on an Interaction Experiment, which specifies the results and details of experimental procedures used to identify biomolecular interactions.
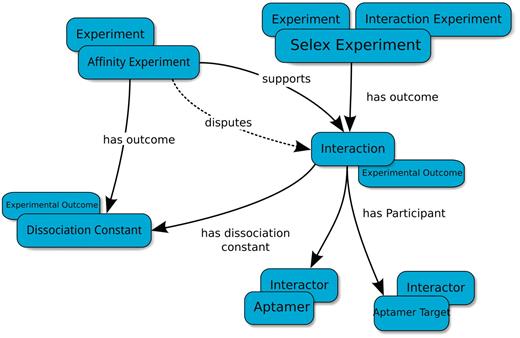
Basic type-relation map used by the Aptamer Base to describe SELEX experiments. The Interaction Experiment type ‘has outcome’ an Interaction. Each Interaction ‘has participant’ at least two Interactors (Aptamer and Aptamer Target). The Affinity Experiment type ‘has outcome’ a Dissociation Constant that either ‘confirms’ or ‘disputes’ an interaction. Blue ellipses denote types and arrows represent properties between topics. Overlapping ellipses represent the multiple types associated with a topic. For more details visit http://aptamerbase.semanticscience.org/?q=node/1.
In the case of aptamers derived from SELEX, the SELEX Experiment captures the SELEX method used to generate the sequences, the partitioning method used to discard non-binding sequences and the recovery method used to isolate the aptamers from the aptamer–target complex. Additional details regarding the SELEX experiment, such as the number of selection rounds, template sequences and details on the selection solution may also be added. The entries for the database were carefully selected based on the methods and results from typical SELEX publications and as suggested by experts in the field (1,2). Interaction Experiments may report one or more Interactions. Each Interaction is related to the participants (Aptamers and Aptamer Targets) of the reported interaction, through the ‘has participant’ property. Interaction Experiments can either ‘confirm’ or ‘dispute’ Interactions under a set of experimental conditions, thus enabling searching for binding and/or non-binding sequences. Finally, the strength of an Interaction is quantified by an Affinity Experiment through a Dissociation Constant. The curated information included SELEX experimental conditions, aptamer sequence(s) and secondary structure predictions, and molecular target details as extracted from each article (Figure 2).
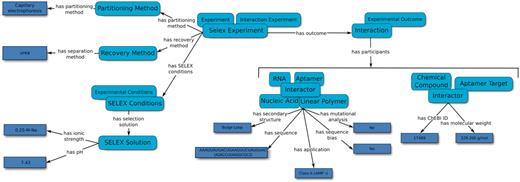
An overview of the relation map used by the Aptamer Base to describe SELEX experiments, the experimental details and the resulting aptamers. Light blue ellipses denote types and arrows represent either properties between topics or between topics and values (dark blue rectangles).
Our data model makes it possible to describe Minimal Aptamers, which are sub-sequences or variants of larger sequences that exhibit binding to a target ligand (Figure 3). Sequences obtained from SELEX are typically at least 30 nucleotides in length (31), but it is widely established that conserved consensus sequences play an important role in biological function. Therefore, a minimal sequence of the full-length aptamer that retains the characteristic function for which the full-length aptamer was originally selected may be identified. The reported minimal aptamers participate in Interactions that are distinct from those of their parent aptamers, thereby permitting the linking of a minimal aptamer–target interaction to a dissociation constant topic that is distinct from that of its parent.
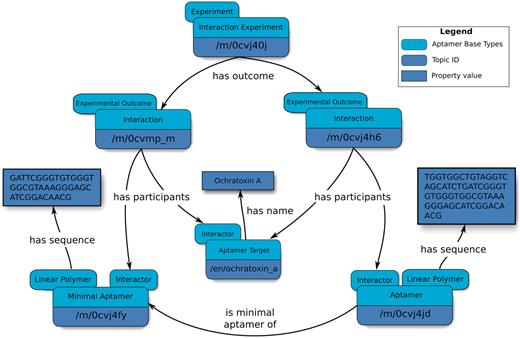
Overview of an abridged relation map describing minimal aptamers. Minimal aptamers are captured in the Aptamer Base by creating a new Interaction for each individual Minimal Aptamer, permitting the description of unique Aptamers can then be associated with the unique Interaction topics.
For every aptamer reported in the Aptamer Base we have also included its secondary structure as predicted by RNAFold (32). The resulting predictions are typed as Predicted Secondary Structure and are associated to Aptamer topics through the Nucleic Acid type by using the ‘has predicted secondary structure’ property. The dot bracket notation, minimum free energy and version of RNAfold are provided for each prediction (e.g.http://www.freebase.com/view/m/0gl4r9m). Given that the ‘has predicted secondary structure’ property is a one-to-many relation, the Aptamer Base could accommodate the incorporation of alternative secondary structure predictions or even tertiary structures made by other prediction programs (33–35).
Curating the aptamer literature
Approximately 3000 articles from the primary scientific literature were identified as potential candidates for inclusion in the Aptamer Base. This collection was created by performing a general search on PubMed for any articles containing the terms ‘SELEX’ or ‘aptamer’ in either the title or abstract. The resulting collection was inspected manually to ensure that only relevant articles, describing a SELEX experiment used to generate aptamers, were assembled for the database entry process. Therefore, all review papers, articles describing applications of previously selected aptamers, articles characterizing or modifying existing aptamers and articles detailing SELEX of in vitro ribozymes were excluded from the final aptamer database collection. The final set of 157 articles published between 1990 and 2006 was curated by six expert graduate students, all of whom have performed experimental research on aptamers or SELEX.
To ensure curation was consistent and accurate, each curator was given the same training in extracting relevant details from aptamer publications. Any curator questions were directed to both of the first authors and our answers were then communicated to all curators. Finally, each entry was verified for completion and accuracy by at least one other curator, and proper typing of the generated Aptamer Base topics was programmatically monitored.
At times, information required in our data model was not present in the primary literature. In some cases this was due to the particular experimental design chosen by the author and in other cases due to incomplete information presented in the research article. To distinguish between this and an incomplete entry by one of the curators, the terms ‘not described’ and ‘not required’ could be selected.
Results
A data summary of the Aptamer Base
The Aptamer Base currently contains information about 676 interactions involving 928 aptamers, 131 targets and 508 dissociation constants organized into 4143 topics, comprising in total approximately 30 000 facts. These facts were added on a weekly basis between September 2010 and July 2011 (http://activity.freebaseapps.com/domain?id=/base/aptamer). More specifically, the Aptamer Base describes interactions involving 328 DNA aptamers and 577 RNA aptamers, with proteins being the most frequently occurring target type (Figure 4).
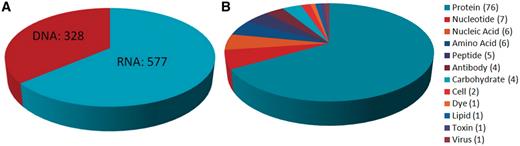
Summary of target types and aptamer types found in the Aptamer Base. (A) Distribution of the over 900 aptamer types described by the Aptamer Base. (B) Distribution of the 131 aptamer targets found in the Aptamer Base.
Accessing and using the Aptamer Base
Searching and browsing data in the Aptamer Base is possible in a number of ways. Firstly, we have created a custom website (http://aptamerbase.semanticscience.org/) which includes a search interface that directly queries the contents of the Aptamer Base (http://aptamer.freebase.com) and presents results via a simple user-friendly search form. Here, the user can narrow his or her search of the almost 1000 aptamer topics by specifying an aptamer type (DNA, RNA or peptide) and/or a target type of interest (see list in Figure 4). Users can also specify a target name in their search by inputting all or part of the target name. The user can choose from a list of available options, which are auto-completed by Freebase Suggest (http://www.freebase.com/docs/suggest). PubMed identifiers can also be specified to limit the search to specific publications. Our custom website also provides users with documentation on using and accessing the data in the Aptamer Base.
Secondly, users have programmatic access to the Aptamer Base through the Freebase application programming interface (API) (http://www.freebase.com/docs/data). The Freebase API allows users to query the Aptamer Base using the Metaweb Query Language (MQL), a powerful tool that allows users to formulate complex queries that are not limited to the type-based searches possible through our search interface. Using the API, queries are serialized as JavaScript Object Notation (JSON) through a Representational State Transfer (REST) interface. A sample MQL query to retrieve all RNA aptamer sequences in the Aptamer Base is shown in Box 1, along with an abridged result set in Table 1.
[{
“id”:null,
“type”:“/base/aptamer/aptamer”,
“a:type”:“/base/aptamer/linear_polymer”,
“b:type”:“/base/aptamer/interactor”,
“c:type”:“/base/aptamer/rna”,
“/base/aptamer/linear_polymer/sequence”:null,
“/base/aptamer/interactor/is_participant_in”:[{
“/base/aptamer/interaction/has_participant”:[{
“d:type”:“/base/aptamer/aptamer_target”,
“name”:null
}]
}]
}]
Abridged list of Aptamer Base topics returned by the query shown in Box 1. The types of the results are not displayed. Topics can be viewed by visiting http://www.freebase.com/view/ [TOPIC_ID]
| Topic ID . | Target Name . | Aptamer Sequence . |
|---|---|---|
| /m/0cvjvjp | Tetracycline | GGCCUAAAACAUACCAGAUUUCGAUCUGGAGAGGUGAAGAAUUCGACCACCUAGGCCGGU |
| /m/0cx03px | Dopamine | GGGAAUUCCGCGUGUGCGCCGCGGAAGACGUUGGAAGGAUAGAUACCUACAACGGGGAAUAUAGAGGCCAGCACAUAGUGAGGCCCUCCUCCCAAGGUCCGUUCGGGAUCCUC |
| /m/0cysc2w | Human epidermal growth factor receptor 3 | CAGCGAAAGUUGCGUAUGGGUCACAUCGCAGGCACAUGUCAUCUGGGCG |
| /m/0czndxr | Human activated protein C | GUGAGACCAGCCGAGUGGUGUCUGGCUAUUCACUGGAGCGUGGGUGGAACCCCUGCGCACUCGUUUGGCUGUCCGGGCCUUCGGGCCGGGAUUAUCUCU |
| Topic ID . | Target Name . | Aptamer Sequence . |
|---|---|---|
| /m/0cvjvjp | Tetracycline | GGCCUAAAACAUACCAGAUUUCGAUCUGGAGAGGUGAAGAAUUCGACCACCUAGGCCGGU |
| /m/0cx03px | Dopamine | GGGAAUUCCGCGUGUGCGCCGCGGAAGACGUUGGAAGGAUAGAUACCUACAACGGGGAAUAUAGAGGCCAGCACAUAGUGAGGCCCUCCUCCCAAGGUCCGUUCGGGAUCCUC |
| /m/0cysc2w | Human epidermal growth factor receptor 3 | CAGCGAAAGUUGCGUAUGGGUCACAUCGCAGGCACAUGUCAUCUGGGCG |
| /m/0czndxr | Human activated protein C | GUGAGACCAGCCGAGUGGUGUCUGGCUAUUCACUGGAGCGUGGGUGGAACCCCUGCGCACUCGUUUGGCUGUCCGGGCCUUCGGGCCGGGAUUAUCUCU |
Abridged list of Aptamer Base topics returned by the query shown in Box 1. The types of the results are not displayed. Topics can be viewed by visiting http://www.freebase.com/view/ [TOPIC_ID]
| Topic ID . | Target Name . | Aptamer Sequence . |
|---|---|---|
| /m/0cvjvjp | Tetracycline | GGCCUAAAACAUACCAGAUUUCGAUCUGGAGAGGUGAAGAAUUCGACCACCUAGGCCGGU |
| /m/0cx03px | Dopamine | GGGAAUUCCGCGUGUGCGCCGCGGAAGACGUUGGAAGGAUAGAUACCUACAACGGGGAAUAUAGAGGCCAGCACAUAGUGAGGCCCUCCUCCCAAGGUCCGUUCGGGAUCCUC |
| /m/0cysc2w | Human epidermal growth factor receptor 3 | CAGCGAAAGUUGCGUAUGGGUCACAUCGCAGGCACAUGUCAUCUGGGCG |
| /m/0czndxr | Human activated protein C | GUGAGACCAGCCGAGUGGUGUCUGGCUAUUCACUGGAGCGUGGGUGGAACCCCUGCGCACUCGUUUGGCUGUCCGGGCCUUCGGGCCGGGAUUAUCUCU |
| Topic ID . | Target Name . | Aptamer Sequence . |
|---|---|---|
| /m/0cvjvjp | Tetracycline | GGCCUAAAACAUACCAGAUUUCGAUCUGGAGAGGUGAAGAAUUCGACCACCUAGGCCGGU |
| /m/0cx03px | Dopamine | GGGAAUUCCGCGUGUGCGCCGCGGAAGACGUUGGAAGGAUAGAUACCUACAACGGGGAAUAUAGAGGCCAGCACAUAGUGAGGCCCUCCUCCCAAGGUCCGUUCGGGAUCCUC |
| /m/0cysc2w | Human epidermal growth factor receptor 3 | CAGCGAAAGUUGCGUAUGGGUCACAUCGCAGGCACAUGUCAUCUGGGCG |
| /m/0czndxr | Human activated protein C | GUGAGACCAGCCGAGUGGUGUCUGGCUAUUCACUGGAGCGUGGGUGGAACCCCUGCGCACUCGUUUGGCUGUCCGGGCCUUCGGGCCGGGAUUAUCUCU |
Data in Freebase is also available as part of the Linking Open Data (LOD) project (36), a community effort that collects data from various open data sources on the Web and publishes and interlinks the different sources using the Resource Description Framework (RDF) (37) in a dereferencable manner. As such, any topic in Freebase and specifically in the Aptamer Base can be retrieved as RDF/XML by using the following URL: http://rdf.freebase.com/rdf/[TOPIC_ID], where TOPIC_ID is the unique identifier given by Freebase to a topic.
Finally, full data dumps of the Aptamer Base are available as tab-delimited files from http://download.freebase.com/datadumps/latest/browse/base/aptamer/.
Discussion
We have described the Aptamer Base, a new collaboratively developed knowledge base motivated by the information needs of aptamer researchers. The Aptamer Base provides detailed, structured information about the experimental conditions under which aptamers were selected and their binding affinity quantified. Our data model provides the required types and properties to capture experimental details of both the partitioning of non-binding sequences from a sequence library and the recovery of binding sequences from aptamer-target complexes in SELEX experiments. The Aptamer Base also describes the pH, temperature, salt concentration and the buffering agent for all SELEX experiments. Providing access to these experimental conditions is crucial in ensuring the reproducibility of the reported experiments (38). This information can be leveraged by aptamer researchers wishing to either reproduce or select new aptamers. For example, researchers attempting to develop aptamers for a specific target can query the Aptamer Base for the conditions of successful SELEX experiments for identical or similar targets.
While earlier efforts at creating databases about aptamers have been documented (24–26), the Aptamer Base provides significant benefits beyond these in several respects. Firstly, it is a part of the larger resource that is Freebase. We have chosen to use Freebase to host the Aptamer Base for a variety of reasons: it provides users with an intuitive interface that readily permits the editing and addition of data, there is a stable community of over 58 000 users that contribute to Freebase on a daily basis, data in Freebase can be queried using the flexible query language MQL, and data stored in this resource is exportable into other data formats (RDF/XML, JSON or text). Secondly, data entry and curation of the Aptamer Base can be undertaken by any registered member of Freebase. This unique feature enables researchers, educators and students within and outside of the aptamer community to contribute and make use of the knowledge in the Aptamer Base. Like other successful open data projects such as Wikipedia, Freebase relies on community collaboration to maintain a complete and accurate dataset. Users can correct or augment facts in the Aptamer Base, but may be suspended if in violation of basic Freebase content creation guidelines (see http://wiki.freebase.com/wiki/Contribution_guidelines). Furthermore, administrators of the Aptamer Base can at any point modify, remove or undo changes to any entry to ensure data quality and consistency To this end, Freebase provides the community with the ability to revert any changes made to the Aptamer Base through the use of Freebase datadumps (http://download.freebase.com/datadumps/), which have complete backups of the data in Freebase for the previous two months. Access to older backups is also available by contacting the Freebase staff.
The Aptamer Base also reuses knowledge that is already in Freebase. Our dataset includes types and topics that have been contributed by the community. For example, we reuse information from Chemistry Commons (http://freebase.com/view/chemistry) to provide links to PubChem and Wikipedia for the majority of aptamer target topics. By choosing to use types and topics already found in Freebase, our curators can save time in the creation of new relevant topics. In addition, curators can ascribe to the community consensus on particular topics and consequently enhance these topics by providing new contextualized knowledge about them. Similarly, the 12 registered members of the Aptamer Base have created types that have been reused by the Freebase staff to annotate 285 Wikipedia articles (http://www.freebase.com/view/base/aptamer/views/rna).
Programmatic access to Freebase data through either the REST interface or their hosted development environment Acre (http://wiki.freebase.com/wiki/Acre) enables its users to create data ‘mashups’ via web applications that use and combine data from different bases in Freebase. These can be combined with external resources to further enhance the knowledge hosted on Freebase. For example, when possible, we provide links to the Chemical Entities of Biological Interest database (ChEBI, available at: http://www.ebi.ac.uk/chebi/) (39), by associating reported small-molecule ligand Interactors with their corresponding ChEBI identifiers. These identifiers can be used to query ChEBI's Simple Object Access Protocol (SOAP) interface to extract further information.
The expandable nature of the data model used by the Aptamer Base has been tailored to provide a core set of the necessary details required to generically reproduce SELEX experiments (38). However, the relatively young stage of aptamer in vitro experiments has inevitably resulted in publications with varying levels of detail. Consequently, the data stored in the Aptamer Base is only as detailed as the publication of origin. Our data model can therefore be used as a common set of requirements for reporting future in vitro aptamer experiments and subsequently expanded to accommodate results of other in vitro sequence evolution experiments that involve biomolecular interactions such as phage display (4) and the development of ribozymes.
Although SELEX identifies putative aptamer sequences, not all have been verified with a binding affinity experiment. As a result, there is always a possibility that these sequences actually have little or no affinity for the target, but have nonetheless been included in an aptamer database. Towards higher quality data, most Aptamer Base Aptamers have been verified by some Affinity Experiment are stated to either bind (‘confirm’) or not to bind to the target (‘dispute’) in question.
With the Aptamer Base, we hope to address the gap that exists in available data resources for in vitro selected sequences. As the biological significance of aptamers becomes better understood, we anticipate that the Aptamer Base will become increasingly important. Rapid advances in structural biology towards the discovery of novel biochemical functions including structure, function and interactions are providing a wealth of information that needs to be accessible by structured queries, whether through simple or programmatic interfaces. The Aptamer Base, through its open framework for accessing and contributing data, promises to not only provide accurate and up to data for aptamer scientists, but also a new resource for bioinformatics research and discovery.
Funding
This work was supported by Natural Sciences and Engineering Research Council Discovery Grants to both MCD and MD. Funding for open access charge: NSERC and Carleton University.
Conflict of interest. None declared.
Acknowledgements
We would like to acknowledge Matthew Chan, Michael Beking, Alexander Wahba and Alison Callahan for their support, useful discussions and technical insight into the creation of the Aptamer Base and writing of this manuscript.
References
Author notes
†These authors contributed equally to this work.



