-
PDF
- Split View
-
Views
-
Cite
Cite
Laura L. Baxter, R. Travis Moreland, Anh-Dao Nguyen, Tyra G. Wolfsberg, William J. Pavan, A curated online resource for SOX10 and pigment cell molecular genetic pathways, Database, Volume 2010, 2010, baq025, https://doi.org/10.1093/database/baq025
Close - Share Icon Share
Abstract
We describe the creation of a specialized web-accessible database named the Pigment Cell Gene Resource, which contains information on the genetic pathways that regulate pigment cell development and function. This manually curated database is comprised of two sections, an annotated literature section and an interactive transcriptional network diagram. Initially, this database focuses on the transcription factor SOX10, which has essential roles in pigment cell development and function, but the database has been designed with the capacity to expand in the future, allowing inclusion of many more pigmentation genes.
Database URL:http://research.nhgri.nih.gov/pigment_cell/
Introduction
Biomedical research is entering a phase of discovery that seeks to incorporate genome-wide and cell system-wide understanding of multiple aspects of the pathways at work in biological systems (1), and expanded resources are needed to facilitate comprehension of these broad research approaches. These resources include centralized web-accessible databases, which can be useful when tailored to a specific research field. Examples include databases for mouse embryonic development (EMAGE mouse embryo gene expression database, http://www.emouseatlas.org/emage), model organisms (the Zebrafish Model Organism database (ZFIN), http://zfin.org/; Mouse Genome Informatics (MGI), http://www.informatics.jax.org/) and specific human disorders (the Inherited Peripheral Neuropathies Database, http://www.molgen.ua.ac.be/CMTMutations/; the Deafness Gene Mutation Database, http://hearing.harvard.edu/db/genelist.htm).
One major focus in the field of pigment cell biology is neural crest-derived pigment cells, termed melanocytes. Melanocytes produce melanin pigment and are primarily located in skin and hair follicles; additionally, populations of melanocytes are found in the inner ear, the eye (choroid, iris, ciliary body and harderian gland) and the leptomeninges of the brain. Melanocytes give coloration to skin and hair, provide protection from solar exposure by increasing melanin pigment in response to ultraviolet radiation, and when malignantly transformed give rise to melanoma, an aggressive cancer of increasing incidence (2,3). Additional pigment cells are found in the retinal pigment epithelium (RPE), an evolutionally conserved pigmented structure of the eye that is essential for normal eye development and vision (4). Unlike neural crest-derived melanocytes, RPE cells develop from the optic neuroepithelium, but RPE cells and melanocytes show overlapping yet non-identical gene expression patterns (5,6). Similarly, cellular pathways essential for melanocyte development and function often overlap with those involved in melanoma progression (7,8).
Historically, pigment cell biology has been at the forefront of genetic discovery, as early understanding of genetic inheritance and linkage was facilitated by analysis of mice that had been selectively bred for various coat colors and patterns for aesthetic reasons. The cloning of the genetic mutations associated with mouse coat color variants revealed not only the genetics of pigment cell biology but also models for human disease and the genetics underlying pleiotropic diseases affecting multiple cell types, where disruption of melanocyte development/function acted as a visible marker for perturbations in the neural crest as well as other lineages (9–11).
Given the wide-ranging applications of pigment cell research, comprehensive compilation of data on all aspects of pigment cell genetics and biology could expedite advances in pigment cell research fields. Therefore, we have created the Pigment Cell Gene Resource (http://research.nhgri.nih.gov/pigment_cell/). The goal of this database is to provide a central location where published data relevant to pigment cells are curated, summarized succinctly and organized in a manner that is easily referenced by the pigment cell researcher, be they someone new to the field becoming acquainted with the literature or an experienced researcher seeking to establish new correlations among existing data. Because this database is tailored for the pigment cell community, it focuses on data relating directly to pigment cell development, excluding data from non-pigment cell systems.
The Pigment Cell Gene Resource is comprised of two sections: an annotated literature section (Figure 1) and an interactive transcriptional network diagram (Figure 2). Although this database is not yet comprehensive for all pigment cell genes and pathways, the annotated literature section does contain complete and detailed information on the transcription factor SRY (sex determining region Y)-box 10 (SOX10). Extensive illustration of numerous pigment cell genetic pathways is provided in the transcriptional network diagram. Importantly, this database has been designed with the capacity to grow and thus include information on many more genes involved in pigmentation.
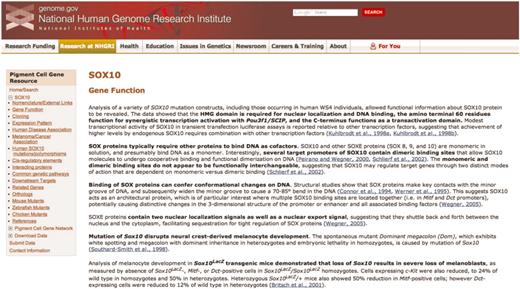
A representative page from the annotated literature section for SOX10 of the Pigment Cell Gene Resource. The navigation sidebar at left includes the 17 literature summary subcategories; each of these categories is located on a separate page. Links to PubMed abstracts for each summarized article are in blue, and bold text indicates the main points of the summary. The navigation sidebar also contains links to access the Pigment Cell Gene Network, to submit data directly and to download data.
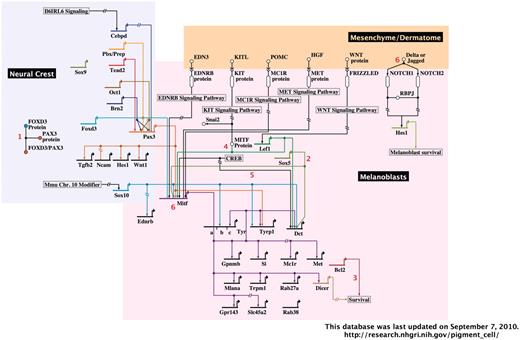
A representative image from the interactive, BioTapestry-generated Pigment Cell Gene Network. The portion of the network containing neural crest and melanoblasts with associated dermatome are shown. Red numbers represent interactive notes, which in the online network become visible by mouse-over. Regions included in the complete diagram are categorized by developmental stage/cell type, as follows: neural crest, melanoblasts and associated dermatome, follicular melanocyte stem cells, follicular/dermal melanocytes and associated keratinocytes and RPE.
Annotated literature summaries
The annotated literature section of the Pigment Cell Gene Resource is a searchable collection of concise summaries of individual peer-reviewed articles, with the main points of the summary in bold text (Figure 1). Links to the associated PubMed abstracts are included. The annotated literature section is categorized by gene; currently presented is SOX10, a transcription factor that is crucial for the development of melanocyte and glial lineages (12–14). Mutation of human SOX10 results in Waardenburg syndrome type 4 and PCWH (peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschsprung disease), disorders that result from abnormal development of neural crest-derived melanocytes, enteric neurons and glia, Schwaan cells and oligodendrocytes (15–17). SOX10 mutations are seen in melanoma as well (18).
Within the annotated literature section, articles are subcategorized using a broad range of topics, including data on both normal and disease states from multiple species. Papers are included in multiple categories when relevant. For example, four separate entries for SOX10 annotate the work of one paper by Britsch et al. (12), because in this paper a new mouse model was created (Sox10LacZ gene insertion), SOX10 function in the survival of developing melanocytes (melanoblasts) was confirmed, Dopachrome tautomerase was shown to be a downstream target of SOX10, and new details of Sox10 expression were revealed. Also, throughout the annotated literature section subcategories are links to SOX10-specific data at relevant sites, including MGI, ZFIN, the University of California Santa Cruz genome bioinformatics browser and the National Center for Biotechnology Information (NCBI) Gene, GenBank and SNP databases. Of note, the ‘Human SOX10 mutations/polymorphisms’ section includes a diagram (Figure 3) and table documenting all published mutations/polymorphisms within the coding region of SOX10, along with their associated phenotypes.
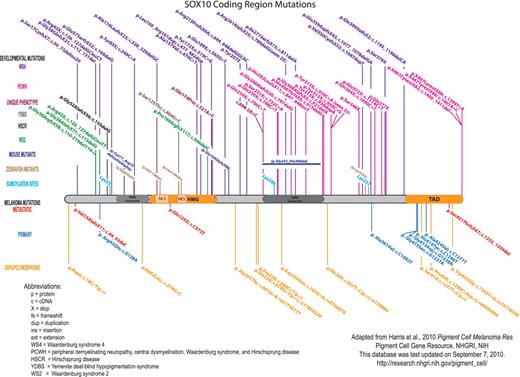
Published SOX10 coding region mutations and polymorphisms. For clarity, the mutations are color-coded based upon clinical phenotype/disorder. Human SOX10 developmental mutations are shown above the protein, and melanoma mutations and polymorphisms are shown below the protein. The orthologous changes to published mouse and zebrafish coding mutations are also shown. SOX10 nucleotide/amino acid numbering correlates with NP_008872. Mutation nomenclature is per Human Genome Variation Society guidelines (http://www.hgvs.org/mutnomen/).
Pigment cell gene network diagram
The second section of this database is comprised of the Pigment Cell Gene Network, an interactive diagram of experimentally verified data describing the gene regulatory networks of melanoblasts, dermal melanocytes, melanocyte stem cells and RPE cells (Figure 2). This diagram was generated using the freely available BioTapestry software (http://www.biotapestry.org/), which allows modeling of the genetic regulation of developmental networks in an easily discernable format (19–21). The use of software designed for gene regulatory network representation is especially applicable given the predicted role of transcription factor cascades in the development and specification of the RPE (5) and neural crest-derived lineages such as melanocytes (8,22–24). BioTapestry’s format of network illustration captures both details and global patterns of expression. For example, it allows visualization of promoter binding site order and thus cis-regulatory information, yet also allows large transcriptional cascades to be readily distinguished by temporal and spatial differences.
The Pigment Cell Gene Network is subdivided by pigment cell type and developmental stage. Thus at a glance, the known interactions that are present in developing melanoblasts can be discerned from those that arise in dermal melanocytes or melanocyte stem cells. Each illustrated gene interaction is annotated with the supporting primary literature, which is accessible by control-clicking (Mac)/right-clicking (PC) on the gene of interest, then selecting ‘Experimental data’ from the menu that appears. Additional useful interactive features on this menu include the ability to highlight pathways of interest, transcriptional sources or targets, or all usages of a gene in the diagram. Additional details regarding cell type, gene interactions, mRNA/protein expression or contradictory data are included in notes (indicated by red numerals; the note text becomes visible in a lower window by mouse-over).
Because the Pigment Cell Gene Network does not currently include all genes/interactions present in pigment cells, it will be expanded in the future. In addition to incorporating more detailed descriptions of current data, future expansions could illustrate spatio-temporal expression. BioTapestry software allows individual gene pathways to be highlighted in conjunction with specific developmental timepoints and locations, and can include time course data and quantitative PCR results. Thus, as precise gene expression patterns are defined during various stages of melanoblast development/migration, melanocyte stem cell regeneration or RPE formation, they can be represented in an unambiguous format.
The Pigment Cell Gene Network currently incorporates data from normal tissue in mammalian systems, thus excluding melanoma as well as zebrafish, xenopus and chick model systems. However, future network expansions could easily include melanoma data and the similarities or differences in the pigmentation gene networks of various organisms. Of note, useful maps for melanoma pathways can currently be found in the Melanoma Molecular Map Project (http://www.mmmp.org) and the KEGG pathway database (http://www.genome.jp/kegg/pathway/hsa/hsa05218.html).
User interface
The web interface for the Pigment Cell Gene Resource was developed using CGI and is hosted on an Apache web server (version 2.0.59) running Linux (Fedora Core 5). The web interface was developed using several programming languages, including: the search function in Perl and CGI; the web front-end in SSI, HTML and Template Toolkit; and the sidebar tree and data download function in Java Script.
With our primary target being the wet-lab biologist, the main design focus of the user interface was ease-of-navigation. The user has the option of directly exploring the Pigment Cell Gene Resource via topic-specific hyperlinks or by entering a keyword search term in the Search box. The search function was designed to return query hits to the search page organized as gene-centric subcategorized topics linked to the appropriate pages. Clicking on any of the returned hits will display the entire list of annotated literature summaries for that particular subcategory.
The navigation sidebar allows the user to peruse the database for a specific pigment cell gene or network, to submit data directly and to download data (Figure 1). The Download Data link is a function that allows the user to generate and save a PDF file of the current display. Optionally, a user may download the entire data set from the Pigment Cell Gene Resource by clicking on the Download Data link from the Home page. The complete data set, organized as gene-centric subcategorized topics, is saved as a Zip archive of individual PDF files. The Submit Data link provides a data upload mechanism for pigment cell researchers to submit published data related to pigment cells and melanoma. Published data are uploaded by selecting a gene of interest and an appropriate data category (e.g. Gene Function), then providing a brief descriptive data summary and a primary literature citation or PubMed identifier. All user-submitted data are reviewed internally prior to full inclusion in the Pigment Cell Gene Resource. The Pigment Cell Gene Network diagram is also accessible from the sidebar; users launch a web-based application using Java Web Start by clicking on the Network Diagram and the subsequent Network Viewer links.
Useful database applications
By creating the Pigment Cell Gene Resource, we sought to fill a need in the pigment cell field that in our opinion was only partially addressed by previously existing databases. Although quite useful, these other databases are neither centralized nor complete with regards to pigment cell biology. For example, the NCBI resource Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim) provides detailed literature summaries and includes some useful details on human developmental disorders and mutations; however, in the case of SOX10 it is incomplete, lacking data on melanoma, animal models, cis-regulatory regions, downstream targets or interacting proteins—all of which are included in the literature section of the Pigment Cell Gene Resource. The International Federation of Pigment Cell Societies’ coat color gene database (http://www.espcr.org/micemut/) is an outstanding resource, documenting cloned and uncloned mutants that affect mouse coat color, and including useful links to genomic and sequence data, gene knockout models and the ZFIN database; however, no primary literature information is included. Large amounts of richly detailed and informative data are available at both NCBI (http://www.ncbi.nlm.nih.gov/) and MGI, but here a researcher must independently parse out data of interest. For example, an NCBI PubMed database search using the terms ‘SOX10’ and ‘melanoma’ retrieves 32 abstracts (7 September 2010, date last accessed), thus necessitating access to 33 different web pages and independent assessment of each article’s content; in contrast, accessing the melanoma/cancer association page of the Pigment Cell Gene Resource immediately retrieves summaries of the 13 most relevant primary research articles. Of note, a human mutation database for all Waardenburg syndrome-associated mutations was recently published, giving excellent documentation of SOX10 mutations and associated clinical phenotypes, but lacking detailed SOX10 literature summaries [http://grenada.lumc.nl/LOVD2/WS/; (25)]; since the Pigment Cell Gene Resource does not contain similarly extensive clinical data, the two databases are complementary.
In regards to the Pigment Cell Gene Network, its key advantages are that it is freely available, annotated with primary literature supporting each illustrated interaction and subdivided by pigment cell type and developmental stage. As a comparison, commercially available systems, such as those offered by Ingenuity (http://www.ingenuity.com/) and GeneGo (http://www.genego.com/), are also interactive, annotated and extremely useful for novel pathway discovery and analysis, especially from large data sets obtained from high-throughput screens. However, they require payment for usage, and also produce networks in a high-throughput fashion, so that analysis of a specific cell type and/or developmental stage typically requires independent filtering and cross-referencing of the primary literature by the researcher.
Future considerations
As with many resources, there are inherent limitations to this database. First, it is dependent on timely manual curation of the rapidly growing body of data describing melanocyte genetic pathways to maintain its relevance. Therefore, this database will be updated at least twice each year. Importantly, the Pigment Cell Gene Resource provides an established framework that will easily support future expansion to include many other genes essential for pigment cell development and function. To facilitate this expansion, we will welcome the participation of other pigment cell researchers through comments/corrections, differences in interpretation and direct contributions in their fields of expertise (via the Submit Data link included at the Pigment Cell Gene Resource).
Secondly, the purposeful selection of articles relevant to melanocytes, RPE and melanoma will at times exclude data that, while seemingly peripheral from the curator’s perspective, may be relevant to a lab with a different focus. For example, the data on SOX10 regulation of glia development in the reference described above (12) are excluded from the Pigment Cell Gene Resource. Broader expansion of this database to include other cell types with related gene regulatory pathways may be warranted. Another concern is that the BioTapestry software used for the Pigment Cell Gene Network is highly useful for the visualization of gene regulatory networks, but is more cumbersome for the illustration of detailed biochemical interactions that occur in the pigment cell, such as receptor-ligand binding, phosphorylation, subcellular localization changes or enzyme kinetics. In response to this problem, however, all data within the Pigment Cell Gene Network diagram are readily exported in Simple Interchange Format (SIF) or Systems Biology MarkUp Language (SMBL) format, so that the existing network data can be manipulated using the numerous network software applications that employ this language (see http://sbml.org/).
In conclusion, we anticipate that the Pigment Cell Gene Resource will help the field of pigment cell biology gain a better understanding of the many systems and pathways that function in pigment cells, given its free access, centralization and anticipated growth to include many more genes in the future.
Funding
Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Conflict of interest. None declared.



