-
PDF
- Split View
-
Views
-
Cite
Cite
Benjamin M. Good, Erik L. Clarke, Salvatore Loguercio, Andrew I. Su, Building a biomedical semantic network in Wikipedia with Semantic Wiki Links, Database, Volume 2012, 2012, bar060, https://doi.org/10.1093/database/bar060
Close - Share Icon Share
Abstract
Wikipedia is increasingly used as a platform for collaborative data curation, but its current technical implementation has significant limitations that hinder its use in biocuration applications. Specifically, while editors can easily link between two articles in Wikipedia to indicate a relationship, there is no way to indicate the nature of that relationship in a way that is computationally accessible to the system or to external developers. For example, in addition to noting a relationship between a gene and a disease, it would be useful to differentiate the cases where genetic mutation or altered expression causes the disease. Here, we introduce a straightforward method that allows Wikipedia editors to embed computable semantic relations directly in the context of current Wikipedia articles. In addition, we demonstrate two novel applications enabled by the presence of these new relationships. The first is a dynamically generated information box that can be rendered on all semantically enhanced Wikipedia articles. The second is a prototype gene annotation system that draws its content from the gene-centric articles on Wikipedia and exposes the new semantic relationships to enable previously impossible, user-defined queries.
Database URL:http://en.wikipedia.org/wiki/Portal:Gene_Wiki
Introduction
Faced with ever expanding amounts of data to process and decreasing budgets, the biocuration community is exploring ways to reduce costs and expand capacity. One avenue of particular interest is the possibility of enabling voluntary community participation in the curation process through open data systems such as wikis (1, 2).
While there are many technical approaches to building wiki software, the one component that is crucial to success is a large, motivated community of users and editors. Generating such a community from scratch is a difficult, complex process that is not well understood and often does not succeed. Because of this challenge, several groups decided to focus their efforts in the context of a community that is already large and active—Wikipedia (3, 4). Rather than building a novel wiki and then working to attract users, these initiatives started with a very large user base and worked to help these users expand and improve the content of the articles relevant to their initiatives. This choice has largely resulted in successes in terms of high quality, volunteer-based content production (5, 6); however, operating in the context of Wikipedia entails a lack of control of the implementation and thus limits what can technically be achieved.
A particular weakness of Wikipedia with respect to biocuration activities is that it is not designed to support the production of structured data. For example, one relevant objective might be to produce a list of all the genes related to a given biological process and also to a particular disease. Since there is no query system in Wikipedia, such lists can only be assembled manually—literally by writing them into a ‘list page’ that must be updated by hand. When the relationships between concepts are structured, for example in a database, it becomes trivial to produce such lists through dynamic queries. The challenge we are faced with is thus to enable the inclusion of structured data in the context of Wikipedia and to do so without the power to change its technical implementation.
Links
In a wiki or any other hypertext system, navigation is made possible through the use of hyperlinks that allow users to jump directly from one document to another related document. In MediaWiki, the engine that powers Wikipedia, the ‘WikiText’ syntax allows adding a hyperlink by enclosing the text of the title of the target page in square brackets. For example, if an editor wanted to add a hyperlink from the article about the 5-HT1A receptor to the article about the biological process of vasodilation, they would simply enclose the appropriate word with double-square brackets (‘[[vasodilation]]’) directly in the text of the 5-HT1A receptor article. When rendered in a Web browser, the 5-HT1A receptor article has a clear hyperlink that users can click to navigate to the vasodilation article.
In addition to aiding navigation for users, hyperlinks may implicitly indicate meaningful relationships between concepts. Where hypertext documents are principally about a particular concept, as is the case in Wikipedia, links between documents provide a loose indication that the concepts represented by those documents are related. In the example above, we may infer the possibility that the 5-HT1A receptor protein plays a role in the process of vasodilation based simply on the presence of the hyperlink connecting the two concepts (7). However, such inferences are imprecise. In the best case, the nature of the relationship between the concepts is undefined (e.g. there is no indication how the 5-HT1A receptor is related to vasodilation) and in the worst case it is non-existent (e.g. the link appeared in the context of a sentence that was not about the 5-HT1A receptor).
Semantic Links make it possible for editors to explicitly specify the meaning of the links in a hypertext document. This construct is the foundation of the Semantic Media Wiki (SMW) system, an extension of the basic MediaWiki framework intended originally to enable the creation of a ‘Semantic Wikipedia’ (8, 9). Within an SMW system, an editor can specify the nature of the relationship between concepts represented on the wiki and the system can then use that additional information to improve the organization of the site. For example, an editor can state that San Diego is a city in California and the system can then use this information to respond to queries like ‘list all cities in California’. Without the addition of the ‘city in’ relationship, such a query would be extremely difficult if not impossible to answer.
While independent wikis, like SNPedia (10), use the SMW extension to more effectively organize the information contained in them, Wikipedia has not yet taken this step. Based on the infrastructure provided by the current Wikipedia platform, it is not possible for an editor to explicitly state the relationships that hold between two entities in a way that can be interpreted without the need for natural language processing. We introduced the Semantic Wiki Link (SWL) template into Wikipedia as a partial solution for encoding, but not querying, semantic content.
Semantic Wiki Links
An SWL is a hyperlink on Wikipedia that allows the editor to explicitly specify the type of relationship between the concept described on the page being edited and the concept that is being linked to (http://en.wikipedia.org/wiki/Template:SWL). These SWLs are implemented using MediaWiki templates. For example, if an editor is editing the page for the gene phospholamban, and wants to specify that its protein product is phosphorylated by protein kinase A, they can indicate this by replacing the normal link [[Protein Kinase A]] with an SWL (Figure 1).
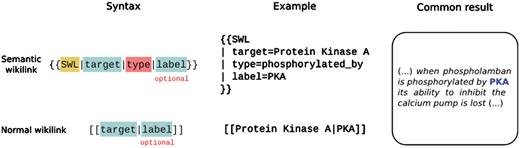
WikiText syntax for Semantic Wiki Link as compared with a normal link.
The MediaWiki system translates the WikiText used by editors into standard HTML understood by Web browsers. During this translation process, WikiLinks like [[Protein kinase A]] become standard HTML hyperlinks like <a href=“/wiki/Protein_kinase_A”>Protein kinase A</a>. SWLs are translated into the following structured HTML (modeled after the ‘microformat’ pattern (microformats.org):
<span class=“swl”>
<span class=“Phosphorylated_by”>
<a href=“/wiki/Protein_kinase_A”>
Protein Kinase A</a>
</span>
</span>
The first class attribute ‘swl’ identifies the presence of a SWL. The class attribute of the next child span element contains a descriptor of the nature of the link encoded (in this case ‘phosphorylated by’). The HTML hyperlink remains unchanged. When an article containing SWLs is rendered in Wikipedia, the links operate in the same way as other links, hence they do not disrupt any existing functionality. But, since SWLs are consistently structured this way, it is possible to write programs that can automatically parse and make use of the information that they encode.
Structuring semantic relationships
Whenever a SWL is inserted into a Wikipedia article, the article containing it is assigned to a Wikipedia category indicating the relationship type employed (e.g. ‘phosphorylated by’). The relationship type category provides a logical grouping (e.g. the things that are phosphorylated by something) and, importantly, a place to define the meaning of the relationship. Meaning is defined in text on the category article, by linking the category to broader categories in the existing Wikipedia category hierarchy and through external links to terms from ontologies on the Semantic Web (Figure 2). Since all SWL link types are visible and editable just like any other Wikipedia category, editors can collaborate on the development of what amounts to a simple ontology of relationship types.
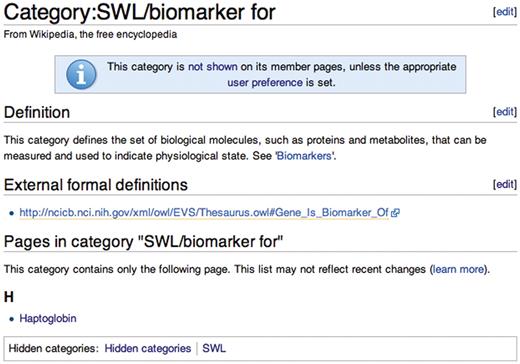
Wikipedia category page for the ‘biomarker for’ relationship type.
Applications
Because Wikipedia is not yet capable of processing semantic relations, SWLs are currently only useful for representing the semantic nature of relationships. To actually make use of the encoded semantic relations, additional tools are necessary. For example, any programmer can now write computer programs to parse Wikipedia content for SWLs and import them into third-party tools (e.g. Cytoscape, triplestores, etc.)
As a proof of concept, we introduce two examples of tools that take advantage of SWLs encoded within Wikipedia. First, we developed a userscript that provides enhanced browsing of Wikipedia pages bearing SWLs. Second, we created another version of the Gene Wiki (4), called ‘Gene Wiki+’, that contains content drawn from Wikipedia but is hosted in a semantically aware environment.
Wikipedia userscripts
Userscripts are javascript programs that can be installed at the discretion of the user to alter the presentation of Web pages. [See (11, 12) for examples in the life sciences.] Wikipedia allows users to customize their user experience by installing userscripts. Commonly used userscripts are included in a set of default scripts that can be activated at the user's discretion as part of their account configuration. It is also straightforward for users to activate scripts authored by other users.
We provide two Wikipedia userscripts, one that uses embedded SWLs to enhance the presentation of the articles where they occur and another to ease the process of inserting them when editing an article. The former generates a tab that, when clicked, renders an infobox displaying all of the SWLs discovered on the page (Figure 3). This table provides a rapid summary of the key attributes of the concept represented by the page (e.g. a gene) and allows the user to quickly navigate to linked articles and to the relationship types used in the SWLs. The other script adds a button to the editing toolbox that helps to produce a correctly formatted SWL (Figure 4).
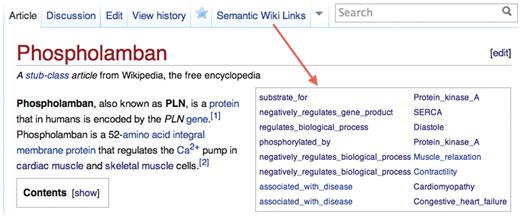
The infobox on the article for Phospholamban generated dynamically with the SWL_infobox user script.
Editing button for help inserting SWLs into Wikipedia articles.
To install these scripts:
Create an account on Wikipedia.
Edit your user page (e.g. http://en.wikipedia.org/wiki/User:yourusernamehere).
Add a sub page that will contain your personal userscripts by inserting: [[/common.js]] on your user page, saving the page, clicking on the resultant red link, and choosing ‘create’.
Edit the common.js page to import the SWL scripts by adding: //SWL editor importScript('User:Genewikiplus/swl_editor.js'); //SWL renderer importScript('User:Genewikiplus/SWL_infobox.js').
Navigate to an SWL-enhanced page such as http://en.wikipedia.org/wiki/Phospholamban
Gene Wiki+
The Gene Wiki is a collection of more than 10 400 Wikipedia articles about human genes (6). We created the Gene Wiki+ application to enable users to execute queries that utilize the semantic relationships encoded in SWLs on these specific articles. Queries such as ‘list the genes that are associated with cancer and are involved in signal transduction’ demonstrate the value of encoding semantic relationships. For each Gene Wiki article, the Gene Wiki+ system transfers the SWL-derived relationships, as well as the rest of the article content, into a separate MediaWiki installation that has been enhanced with the Semantic MediaWiki extension (9).
During the transfer process, the system detects SWL templates in relevant Wikipedia articles and converts them to semantic links as they are represented in the Semantic Media Wiki framework. For example, the article on adenosine deaminase contains the following SWL:
{{SWL | target=hemolytic anemia | type=overexpression_results_in}}
When the article is transferred to Gene Wiki+, the system translates this to the Semantic MediaWiki equivalent:
[[overexpression_results_in::hemolytic anemia]]
This translation has two immediate consequences in the context of Gene Wiki+. First, the semantic link is visible through the ‘browse properties’ feature when viewing either the adenosine deaminase article or the article on hemolytic anemia (Figure 5). Second, the relationship can be used in queries such as ‘list all the genes whose overexpression results in hemolytic anemia’. These semantic features are enabled as default behaviors of the Semantic Media Wiki system and hence accessible through the Gene Wiki+ instance. In addition, the semantic relationship type for the SWL is brought over and encoded as a Semantic Media Wiki Property. This allows Gene Wiki+ users to view its textual definition and to navigate to related properties within external vocabularies.
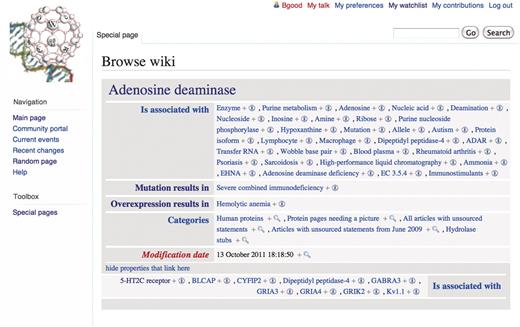
Semantic Media Wiki browse by properties feature accessible in Gene Wiki+. Displaying semantic content from article about adenosine deaminase.
As illustrated in Figure 5, the Gene Wiki+ system contains semantic links translated from SWLs in Wikipedia such as ‘mutation results in’ and ‘overexpression results in’, but it also contains a large number of ‘is associated with’ relationships. The system generates these loosely typed relationships whenever a standard (i.e. untyped) hyperlink is detected in the source article on Wikipedia. While it will take time for SWLs to be inserted into Wikipedia, these simple associations enable interesting queries immediately. For example, a simple hyperlink to the article on phosphorylation is translated to [[is_associated_with::phosphorylation]]. Genes that code for kinases, for instance, nearly all include this particular link; translating it to a semantic link allows us to search for all genes that are associated with phosphorylation. The semantic query can then be expanded to find genes that are involved with phosphorylation and also in cancer, and so forth. These queries will become more powerful as these generic ‘is associated with’ relationships are made more precise (‘catalyzes’ for kinases, ‘catalyzes removal’ for phosphatases, etc.).
Aside from its use in direct association queries, the ‘is_associated_with’ property also forms the root of the property hierarchy used in Gene Wiki+. This means that queries for X where X ‘is_associated_with’ Y will return results where X ‘some more specific relationship’ Y. Figure 6 illustrates how simple associations and more specific semantic relationships can be blended in a Gene Wiki+ query. The table returned by the query lists all genes related somehow to hemolytic anemia. In addition, it presents specific semantic relationships, such as ‘overexpression results in’ or ‘biomarker for’ that link each gene to hemolytic anemia and to other diseases.
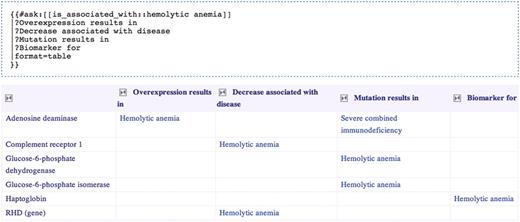
Selecting genes related to hemolytic anemia and exposing the nature of those relationships. The Semantic MediaWiki query is presented above the results table that it generates.
The process of transferring content from Wikipedia to Gene Wiki+ runs continuously such that all changes to relevant articles on Wikipedia are brought over to the Gene Wiki+ in near real time. This makes it possible for users to edit Wikipedia articles with SWLs and then immediately see how those changes impact queries in Gene Wiki+. The articles brought over from Wikipedia are not editable on the Gene Wiki+; any changes should be made directly on the original Wikipedia article.
The software that manages the transfer of data between Wikipedia and Gene Wiki+ is general enough to support the mirroring of any MediaWiki instance from another and is available at (http://genewikiplus.org/wiki/MediaWikiSync).
Gene Wiki+ and the Semantic Web
In addition to providing novel functionality for end users, the Gene Wiki+ system provides access to its content for developers in the form of a Resource Description Framework (RDF) export (http://genewikiplus.org/wiki/GeneWiki:Data). This export allows developers to make use of the growing number of tools for processing RDF content to perform queries over the encoded relationships and to integrate the exported data with their own data.
To facilitate integration with other Linked Data resources, all Gene Wiki+ articles are annotated with their equivalents in DBpedia (13). DBpedia is a Web-accessible RDF database constructed automatically by extracting data from the few consistently structured parts of Wikipedia pages such as categories and ‘infoboxes’. Infoboxes display facts as key-value pairs rendered in a table typically visible on the upper right of Wikipedia articles. For example, there is a taxonomy infobox that contains data about the scientific classification of organisms (e.g. kingdom, phylum, etc.). Where available, such data is extracted automatically and represented in the DBpedia system. The DBpedia system makes no attempt to extract data from the hypertext of Wikipedia articles and, as such, is entirely complementary to the data represented in embedded SWLs. Mapping Gene Wiki+ to DBpedia provides the opportunity to very easily integrate the information derived from SWLs embedded in article text with what structured data does exist on Wikipedia. In addition, DBPedia is a central point for ontology term mapping for the Semantic Web and, as a result, integration with it begins the process of integration with the many other knowledge bases that also map their concepts to it.
Aside from DBpedia integration, the Gene Wiki+ also produces equivalency links between relationship types in SWLs and properties in external ontologies. This is achieved by processing SWLs that appear on relationship type pages in Wikipedia and detecting when they contain the ‘equivalent’ relationship. For example, the relationship type page for ‘biomarker for’ contains the SWL:
{{SWL | type=equivalent
| target=http://ncicb.nci.nih.gov/xml/owl/EVS/ Thesaurus.owl#Gene_Is_Biomarker_Of
}}
In the RDF generated by the Gene Wiki+, this is translated to establish an ‘equivalentProperty’ link between the Gene Wiki+ biomarker property and its equivalent in the National Cancer Institute (NCI) Thesaurus. Establishing such mappings facilitates the process of integrating RDF-based data assembled at multiple locations—in this case it would help to allow data from the Gene Wiki+ to be aggregated with data from the NCI.
Gene Wiki+ future
The Gene Wiki+ is currently a one-way mirror of the Gene Wiki content on Wikipedia that can make use of the semantic relationships encoded by SWLs. Using the code developed to handle the mirroring, we have also begun to incorporate related content from SNPedia, a semantic wiki devoted to information linking single nucleotide polymorphisms to human disease (14). In future work, we may incorporate other data sources and, if so, we will focus primarily on other wiki-based resources that are not currently well represented on the Semantic Web.
Discussion
Wikipedia hosts the largest and most prolific community of producers of textual knowledge on the Web. While the volume of knowledge represented within Wikipedia is vast, it is difficult to extract that knowledge in the form of structured data for computation and analysis. Many users within the Wikipedia community have noted the huge potential benefit of integrating a fully semantically aware infrastructure, but previous proposals have involved technical changes that the governing body of Wikipedia has resisted (8).
Here, we implement a partial solution by decoupling the encoding of semantic relationships from the querying and utilization of those relationships. We provide a mechanism for the massive Wikipedia community to participate in the process of assembling not just a textual resource, but a far more powerful structured knowledge base. Mining and utilizing that knowledge base can then be performed using third-party tools without requiring changes to Wikipedia itself.
Of course, this approach is not without its challenges. Since anyone can create a SWL and there is no way to enforce the use of a particular set of relationship types, it is certain that there will be some inconsistency in how they are used. For example, it is possible that one editor might use the relationship type ‘phosphorylated by’ while another editor might insert ‘phosphate group added by’. Such inconsistencies disrupt semantic queries made over aggregated content such as those available through the Gene Wiki+. However, this is no different from other aspects of Wikipedia articles. Over time, the Wikipedia community has gradually moved toward consensus regarding the use of categories, when to insert normal WikiLinks, when to insert references and how to format them, how to format articles and even how to style the text in the articles. Since Wikipedia is a continuously changing, social artifact there will always be exceptions to the socially defined rules that have emerged to govern its content, but overall, the basic structures remain remarkably stable. If the Wikipedia community takes up SWLs, we expect the same kind of social consensus to emerge with respect to their use. The community will evolve rules defining which properties should apply to which kinds of entities and will police the articles for adherence to these rules in the same way they do now for other article attributes. The key question is whether the community will in fact buy in to the SWL idea.
Right now, there are only a handful of SWLs active on Wikipedia, all of which have been deposited by our team as demonstrations to seed the process. If the SWL concept succeeds, this number should rapidly increase into the tens of thousands, but for that to happen, the Wikipedia editor community must become involved as semantic link authors. In order to recruit this labor, the value of such work needs to be clearly apparent. Since Wikipedia itself does not process the semantic relationships, external applications need to be developed and promoted that make the value of these contributions clear, thereby providing editors with vital positive feedback. Applications like Gene Wiki+ and the infobox-generating userscript provide some first steps in this direction. These applications demonstrate that adding semantic relationships can substantially enhance users interactions with the knowledge in Wikipedia and can enable the production of novel applications relevant to biocuration activities.
Whether these applications will be enough to motivate the Wikipedia community to accept and use SWLs is an open question. In our early interactions with the editor community, there has been some resistance to the use of the SWL template on the grounds that it makes the WikiText more difficult to edit. However, there have also been some enthusiastic responses from community members who see the potential of the idea. As this project unfolds over time, we will work with the editors to come to a solution that the community can wholeheartedly accept.
Conclusion
This work provides an interface between a massive community of knowledge producers and an emerging ecosystem of semantic technologies. It renders the unparalleled ‘crowd power’ of the Wikipedia community accessible for the work of establishing structured relationships between concepts. In short, SWLs enable the Wikipedia community to directly write to the Semantic Web. In doing so, it substantially enhances Wikipedia's value as a platform for collaborative knowledge synthesis in biology and other domains.
Funding
National Institutes of Health (grant number GM089820). Funding for open access charge: National Institutes of Health.
Conflict of interest. None declared.



