-
PDF
- Split View
-
Views
-
Cite
Cite
Margarida Julià-Sapé, Miguel Lurgi, Mariola Mier, Francesc Estanyol, Xavier Rafael, Ana Paula Candiota, Anna Barceló, Alina García, M. Carmen Martínez-Bisbal, Rubén Ferrer-Luna, Àngel Moreno-Torres, Bernardo Celda, Carles Arús, Strategies for annotation and curation of translational databases: the eTUMOUR project, Database, Volume 2012, 2012, bas035, https://doi.org/10.1093/database/bas035
Close - Share Icon Share
Abstract
The eTUMOUR (eT) multi-centre project gathered in vivo and ex vivo magnetic resonance (MR) data, as well as transcriptomic and clinical information from brain tumour patients, with the purpose of improving the diagnostic and prognostic evaluation of future patients. In order to carry this out, among other work, a database—the eTDB—was developed. In addition to complex permission rules and software and management quality control (QC), it was necessary to develop anonymization, processing and data visualization tools for the data uploaded. It was also necessary to develop sophisticated curation strategies that involved on one hand, dedicated fields for QC-generated meta-data and specialized queries and global permissions for senior curators and on the other, to establish a set of metrics to quantify its contents. The indispensable dataset (ID), completeness and pairedness indices were set. The database contains 1317 cases created as a result of the eT project and 304 from a previous project, INTERPRET. The number of cases fulfilling the ID was 656. Completeness and pairedness were heterogeneous, depending on the data type involved.
Introduction
Brain tumours afflict a larger percentage of the European population as life span increases, and in children over 1 year of age, they are the most common cause of death from disease. Their characterization using magnetic resonance imaging (MRI), such as regional extent, oedema and mass effect, is non-invasive. The achievable Sensitivity and Specificity with radiological analysis of an MRI study vary depending on the tumour type and grade (1), and histopathological analysis of a biopsy sample remains the gold standard for their classification. However, stereotactic brain biopsy has significant risks, with an estimated morbidity of 2.4–3.5% (2, 3) and a death rate of 0.2–0.8% (2, 4). There are situations in which repeated biopsies may not be advisable or practical, as in brain tumours of children or in aged patients in a bad physical condition. Hence, there is a need for improving the non-invasive brain tumour diagnostic and prognostic characterization (5), to improve patient management and treatment.
The eTUMOUR (eT) project (Web Accessible MR Decision Support System for Brain Tumour Diagnosis and Prognosis, incorporating in vivo and ex vivo Genomic and Metabolomic Data) was an EU-funded effort (FP6-2002-LIFESCIHEALTH 503094), carried out from February 2004 to August 2009 (5.5 years), involving 21 partners across Europe and Argentina. The aim of the project was to coordinate European scientific expertise in magnetic resonance spectroscopy (MRS) and genetic analysis to improve tumour classification and provide health care professionals with a validated decision support system (DSS) for non-invasive, MRS-based characterization of brain tumours. The project built upon expertise obtained with the INTERPRET EU project IST-1999-10310 (January 2000–December 2002), which created an MRS-based DSS for tumour diagnosis (6), and an associated database (7), which is still accessible to the medical community. The goal of eT was to bring together the expertise required to study the genomic and metabolomic characteristics, both in vivo and ex vivo of brain tumours, with a multi-centre collaboration to acquire statistically significant data and to investigate how metabolomic and genomic profiles relate to diagnosis or to clinically relevant factors such as survival time and treatment response.
Much of the effort of the project was put in data gathering and database development and annotation. Partners and countries involved in data acquisition are listed in Table 1.
| Partner (acronym) . | City . | Country . |
|---|---|---|
| Universitat Autònoma de Barcelona (UAB) | Cerdanyola del Vallès | Spain |
| Hospital Sant Joan de Déu (HSJD) | Barcelona | Spain |
| University of Birmingham (UB) | Birmingham | UK |
| Fundación Lucha contra las Enfermedades Neurológicas de la Infancia (FLENI) | Buenos Aires | Argentina |
| Cancer Research UK Cambridge Research Institute (CRUK) | Cambridge | UK |
| Deutsche Krebsforschungzentrum Heidelberg (DKFZ) | Heidelberg | Germany |
| Medical University of Lodz (MUL) | Lodz | Poland |
| St Georges’ Hospital and Medical School (SGUL) | London | UK |
| University Medical Centre Nijmegen, Radboud University (RU) | Nijmegen | The Netherlands |
| University of Nottingham (NOTTI) | Nottingham | UK |
| Universidad de Valencia (UVEG) | Valencia | Spain |
| Partner (acronym) . | City . | Country . |
|---|---|---|
| Universitat Autònoma de Barcelona (UAB) | Cerdanyola del Vallès | Spain |
| Hospital Sant Joan de Déu (HSJD) | Barcelona | Spain |
| University of Birmingham (UB) | Birmingham | UK |
| Fundación Lucha contra las Enfermedades Neurológicas de la Infancia (FLENI) | Buenos Aires | Argentina |
| Cancer Research UK Cambridge Research Institute (CRUK) | Cambridge | UK |
| Deutsche Krebsforschungzentrum Heidelberg (DKFZ) | Heidelberg | Germany |
| Medical University of Lodz (MUL) | Lodz | Poland |
| St Georges’ Hospital and Medical School (SGUL) | London | UK |
| University Medical Centre Nijmegen, Radboud University (RU) | Nijmegen | The Netherlands |
| University of Nottingham (NOTTI) | Nottingham | UK |
| Universidad de Valencia (UVEG) | Valencia | Spain |
| Partner (acronym) . | City . | Country . |
|---|---|---|
| Universitat Autònoma de Barcelona (UAB) | Cerdanyola del Vallès | Spain |
| Hospital Sant Joan de Déu (HSJD) | Barcelona | Spain |
| University of Birmingham (UB) | Birmingham | UK |
| Fundación Lucha contra las Enfermedades Neurológicas de la Infancia (FLENI) | Buenos Aires | Argentina |
| Cancer Research UK Cambridge Research Institute (CRUK) | Cambridge | UK |
| Deutsche Krebsforschungzentrum Heidelberg (DKFZ) | Heidelberg | Germany |
| Medical University of Lodz (MUL) | Lodz | Poland |
| St Georges’ Hospital and Medical School (SGUL) | London | UK |
| University Medical Centre Nijmegen, Radboud University (RU) | Nijmegen | The Netherlands |
| University of Nottingham (NOTTI) | Nottingham | UK |
| Universidad de Valencia (UVEG) | Valencia | Spain |
| Partner (acronym) . | City . | Country . |
|---|---|---|
| Universitat Autònoma de Barcelona (UAB) | Cerdanyola del Vallès | Spain |
| Hospital Sant Joan de Déu (HSJD) | Barcelona | Spain |
| University of Birmingham (UB) | Birmingham | UK |
| Fundación Lucha contra las Enfermedades Neurológicas de la Infancia (FLENI) | Buenos Aires | Argentina |
| Cancer Research UK Cambridge Research Institute (CRUK) | Cambridge | UK |
| Deutsche Krebsforschungzentrum Heidelberg (DKFZ) | Heidelberg | Germany |
| Medical University of Lodz (MUL) | Lodz | Poland |
| St Georges’ Hospital and Medical School (SGUL) | London | UK |
| University Medical Centre Nijmegen, Radboud University (RU) | Nijmegen | The Netherlands |
| University of Nottingham (NOTTI) | Nottingham | UK |
| Universidad de Valencia (UVEG) | Valencia | Spain |
In some cases, a single partner could in turn coordinate data acquisition at several collaborating clinical centres, i.e. UAB coordinated Institut de Diagnòstic per la Imatge, Centre Bellvitge (IDI-Bellvitge) and Centre Badalona (IDI-Badalona), IAT/CRC CIM Centre d'Imatge Molecular del Parc de Recerca Biomèdica de Barcelona (PRBB), Centre Diagnòstic Pedralbes (CDP), Hospital Universitari de Bellvitge, Hospital Germans Trias i Pujol and Hospital del Mar. Similarly, UVEG coordinated data gathering at Hospital La Ribera-Alzira, Hospital Quiron Valencia, Instituto Valenciano Oncología, Hospital Clínico Universitario- Valencia, Hospital Universitario Dr. Peset and Hospital Universitario La Fe Valencia.
The database was planned to store, in 5 years, data from about 1000 patients acquired at different countries and with different MR scanners. To achieve this, the project had a devoted data ‘workpackage’ (the term used in European Framework Program jargon), with the following goals:
To discuss and agree the consensus acquisition protocols, and to acquire the corresponding data.
To develop a web-accessible database for standardized data storage, guaranteeing data privacy, confidentiality and authenticity.
To use quality assessment practices to ensure compliance with ethical aspects (patient consent forms, data anonymization and local ethics committee approval).
This article will focus on the description of the methodology followed and the challenges met to develop and annotate the eT database (eTDB).
Challenges determining the functional requirements
The project was set up in a way where the eTDB was the first stage of the project (Figure 1). The database had to be functional and contain good-quality data so that pattern recognition specialists (mathematicians and statisticians) could download good quality datasets for their experiments, which would in turn result in classifiers to be incorporated into the DSS. Functional requirements were obtained from users (clinicians, spectroscopists and molecular biologists) according to what would be an ideal way of organizing and storing the types of information that was being collected, in accordance to the acquisition protocols set up for the project (MR protocols: http://www.etumour.net/index.php?option=com_content&task=category§ionid=26&id=204&Itemid=181; Clinical protocols: http://www.etumour.net/index.php?option=com_content&task=category§ionid=26& id=203&Itemid=183).
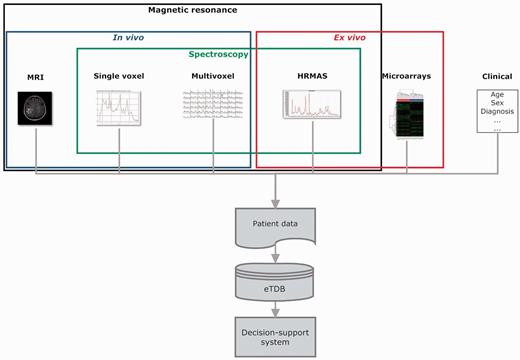
Overview of the eT project with respect to data. The database is built on the concept of patient and each patient has different data types.
Three main challenges were faced. The first one was to agree on the exact way to store all the data collected. The second was that the database must allow expert-based quality control (QC) on the data, to avoid ‘garbage in, garbage out’ in the pattern recognition stage. Mathematicians had the entire responsibility in classifiers and curators were only requested to validate results after the classification studies. In addition, only previous experience on intensive QC was available from INTERPRET (clinical and SV data) but no further automated QC tools were available in 2004, e.g. for high-resolution magic angle spinning (HRMAS). Therefore, the QC had to be a combination of expert-based and automated methods for the different data types. As for the expert-based methods, it was mandatory to incorporate visualization tools for the raw data, because in MRS, raw data signals come in the time domain, i.e. signal intensity versus time, in very specific formats depending on the manufacturer (Supplementary Table S1). In this context, visualization implied that MRS raw data had to be processed by the eTDB, rendering images of processed spectra in the frequency domain, which would be the meta-data on the web-based interface.
The third challenge was first to be able to monitor the progression in the number and quality of the available data throughout the project’s development and second to establish criteria to set up a ‘validated database’ (7), i.e. a ‘quality dataset’, with which further studies could be undertaken, similarly to the concept of inclusion criteria of clinical studies.
Data types
The basic unit in the eTDB is the patient. Five data types were defined for each patient (Figure 1), i.e. clinical, in vivo single voxel (SV) MRS, multi-voxel (MV) MRS, MRI and ex vivo HRMAS and microarray data. The clinical information was mostly text, although representative images from the histopathological slides could also be stored. For the rest of data types, the database contained both the raw data (Supplementary Table S1) and a set of specific meta-data. By meta-data, we mean acquisition parameters, validation-related information and processed MRS data. The first two were text fields that could be automatically extracted from raw data, manually uploaded by curators or automatically calculated by the eTDB. Processed MRS were txt, jpg or pdf files, depending on data types.
The web-based application and user interface
The eTDB is implemented on an Oracle 10 g Database Management System server running on a Linux box (Ubuntu Debian 32-bit). It is based on the three-tier application model, which is composed by three main components: the client (the browser application), the server (Apache Tomcat 5.5.17) and the database (Oracle 10g). The graphical user interface (GUI) developed as the front-end for the eTDB is based on Java Server Pages and JavaBeans (http://java.sun.com/javase/technologies/desktop/javabeans/index.jsp). Additional functionalities, such as file upload or the interface between the GUI and the JavaBeans, were implemented using Java servlets (http://java.sun.com/products/servlet/). All incoming patient data (in vivo MRS and MRI) from the different manufacturers were anonymized or de-identified at the time of MRS and MRI upload with java applets (http://java.sun.com/applets/) within the system’s web front end. Once the file was read and parsed through the applet, the fields of interest (e.g. name, id code, date of birth, sex) found within were removed and a compressed (zip) file was created on the curator’s computer. This compressed file was the raw data subsequently entered into the eTDB. The format, and therefore the structure, of these files is entirely dependent on the manufacturer and data type, and until now there is no standard way of delivering this type of data, despite the DICOM (Digital Imaging and Communication in Medicine) standard having been defined for MRS data since 2002 (http://www.dclunie.com/dicom-status/status.html). For this reason, the anonymization applet within the eTDB possesses an adaptor for each of the formats encountered throughout the development of the project (Supplementary Table S1).
Figure 2 explains the procedure for collecting and implementing the specifications, which was refined through the project’s progress and carried out up to the last quarter of the fifth year, since the complexity of the requirements increased as the project advanced. Regarding users, the ‘data team’ refers to the core team that took over the developments in the database. It included the database manager, who was also the database programmer; the data manager who was also the person communicating both with the users and the database manager and the data workpackage leader, who coordinated the data team.
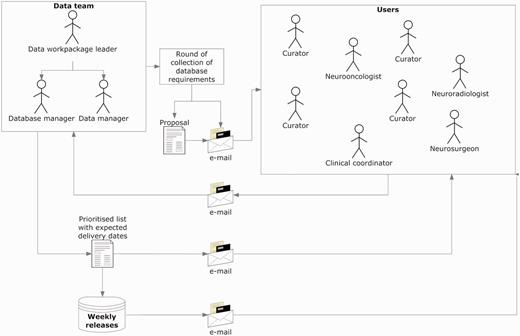
Collecting and implementing specifications. The data manager announced a round of collection of specifications, with a deadline for receiving comments and counterproposals (1–2 weeks). Generally, all requirements were incorporated, except those clearly not feasible. In case of conflicting requirements, the data team prioritized after discussion with the users making the request. Implementations were transparent to users, and when the service had to be interrupted to upload the changes, the database manager sent an announcement to the general mailing list.
User roles are summarized in Figure 3. The other roles allowed in the system were as follows:
Curator: the basic role of the system and therefore was applied to most users. It is basically a data uploader, with edit permissions over the data belonging to the user’s centre, but no permissions to edit data from other centres. As a general rule, it was set to enable viewing the data from all centres.
Senior curator: this role is meant only for the expert spectroscopists (ES) or the clinical coordinator. ES are not allowed to modify clinical data or any type of raw data on any of the cases on the database, and although they still belong to one of the centres registered within the database, ideally they should not be involved in data entry procedures. This role is the only one able to see the QC section of the experiments (clinical or spectroscopic), and the cases and experiments on which the expert is able to edit this section is determined by a set of permissions that are stored inside the database, which are retrieved in real time for adjusting the interface settings and layout.
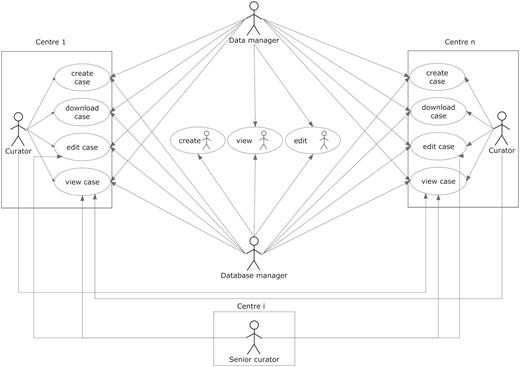
User roles and their permissions. Permissions to perform the different tasks are stored within the database and are based on the centre the user belongs to.
Aside from the roles, the system has also a set of embedded permissions, applicable over any kind of user and has priority over the roles just described. These are: edit, view, create and download and when enabled, will allow a user to edit, view, create and download a case (including all of its sections). They are organized by centres, which means that if a user is given ‘view’ permission over centre ‘A’, he/she will be able to view the data from the cases belonging to centre ‘A’, when originally he/she could only see the data of his/her centre’s cases. A typical curator will have all the four types of permissions over the centre he/she belongs to, although additionally it can have a specific kind of permission over other centres. For example, during the project, those researchers involved in carrying out pattern recognition analysis of the data were able to download data from all cases in order to analyse them, therefore having the ‘download’ permission for all centres in the eTDB.
The eTDB web-based application offers a comprehensive suite of queries to allow any user to search the cases in the database by any field. A set of 21 pre-defined queries, the ‘simple queries’ were designed to fulfil the most common needs of most users with one click. For example, looking for cases with a specific data type from a given centre, entered after a certain date. Or, for each ES, which cases have MRS pending of his/her rating. The ‘advanced queries’ allow users to establish filtering criteria, as well as selecting which fields will be presented in the results table.
Visualization
All raw data are available for download in their original formats, for users with download permissions so that they can process themselves with the software and parameters of their choice, because the specific processing of the MRS can be varied and influences subsequent analysis. However, to complete the expert-based QC, MRS data had to be presented to ES in a processed form. This was achieved by either automated or manual processing which rendered spectroscopic meta-data in the form of images for web display in the eTDB interface. The processing relied on consensus processing parameters, agreed as well in the course of the project.
The in vivo SV MRS data processing was performed with the routines inherited from the INTERPRET project (6), which are called Data Manipulation Software (DMS) (http://gabrmn.uab.es/DMS) (8, 9, 10). This software generates two meta-data (image files) at two different frequency ranges (0–4.5 ppm and 0–12 ppm) for any given spectrum, to facilitate visual analysis through the web-based interface (Supplementary Figure S1). As in the case of the anonymization applet, the DMS had been developed until 2002 and formats posterior to this date cannot be handled by the DMS. A roundabout to solve this was the replication of all meta-data fields to accept manually processed spectra using one of the most common and robust available softwares for spectral processing: jMRUI (Java-based Magnetic Resonance User Interface) (11). The DMS was used as a converter jMRUI → DMS format (10). There were no automated routines for processing MV data, as well as no consensus display format such as the one generated by the DMS, therefore the raw MV data and the meta-information of the processed MV grid had to be uploaded separately in a web browser-readable format, which conditioned a series of additional fields in the interface. HRMAS experiments (Supplementary Figure S2) suffered from the same caveat as MV. The curator had to upload both the raw data and the manually processed images of the spectra into the system. As before, the images must be uploaded in a web image format. The MRI, which are commonly delivered in the DICOM format (http://medical.nema.org/dicom/2004.html), were automatically processed by the database with either the dicom2 application (http://www.barre.nom.fr/medical/dicom2/) or the ImageMagick software suite (http://www.imagemagick.org/script/index.php). This allowed the system to obtain the images in a web-based format, which were in this way, copied to the server’s hard drive and an appropriate pointer was generated inside the database for each of them, which maintained their location on the hard drive for its display on the web application.
In the case of microarrays (Supplementary Figure S3), the web interface does not present graphical visualizations, because raw data were simply uploaded into the system and stored along with the other meta-data that comprised the experiments’ information.
Curation workflow
Curation was divided into two activities: data entry and data validation. A workflow is shown in Figure 4.
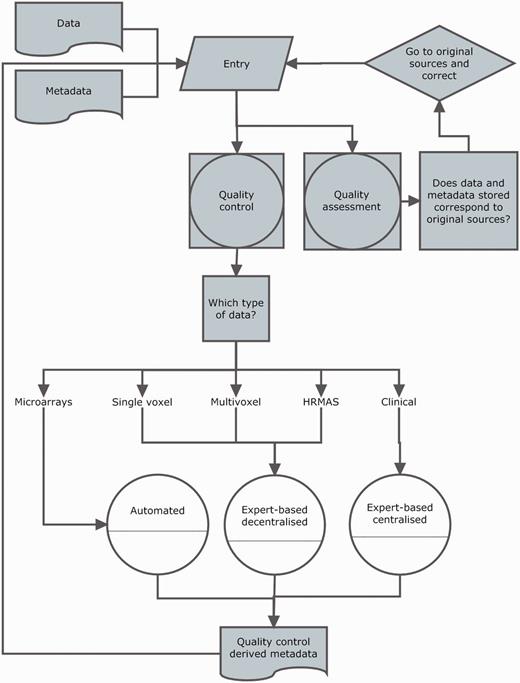
Curation workflow. Diagram showing the data entry, the three types of quality control (white circles), quality assurance and how they relate to different data types and meta-data and to each other.
Data entry was performed by curators, and each centre could have specific personnel with different areas of expertise (microarrays, spectroscopy, radiology). In most cases, the same person that acquired the in vivo data (radiologists) or was responsible of biopsy processing and analysis (HRMAS or microarrays) was the one who performed data entry. With respect to clinical information and in vivo MRS, in most cases, there were support curators (biochemists or biomedical spectroscopists) who performed data entry in collaboration with radiologists, pathologists or neurosurgeons.
From the point of view of the system and its interface, data entry consisted of two main actions performed by the curator: filling out the meta-data text fields and uploading the raw data files. In each of the sections of any given case, there were particular fields to specify the raw data location on the client machine, which were then processed by existing meta-data reading or pre-processing pipelines for their display into the web-based interface.
Validation was set at two levels (Figure 4): (i) QC, in which the purpose was to establish, for each section, if the data entered satisfied the standards (from the instrumental point of view) set by acquisition protocols; (ii) quality assessment, in which the purpose was to establish whether the eTDB entries were the same as source records, i.e. hospital files.
The QC of all spectroscopy data (SV, MV and HRMAS) underwent a three-expert system evaluation, and the general procedure is illustrated in Figure 5. Each ES was assigned to centres different from their own to perform the QC: this was done to ensure that no expert had seen the data being evaluated before and that he/she had no preference on accepting the data for reasons other than its validity and integrity and that he/she had not been involved in the process of acquisition or uploading of the evaluated data. The assignment of centres for evaluation was stored into the database for reference by the system, which consulted this information in real time every instance an ES logged into the system.
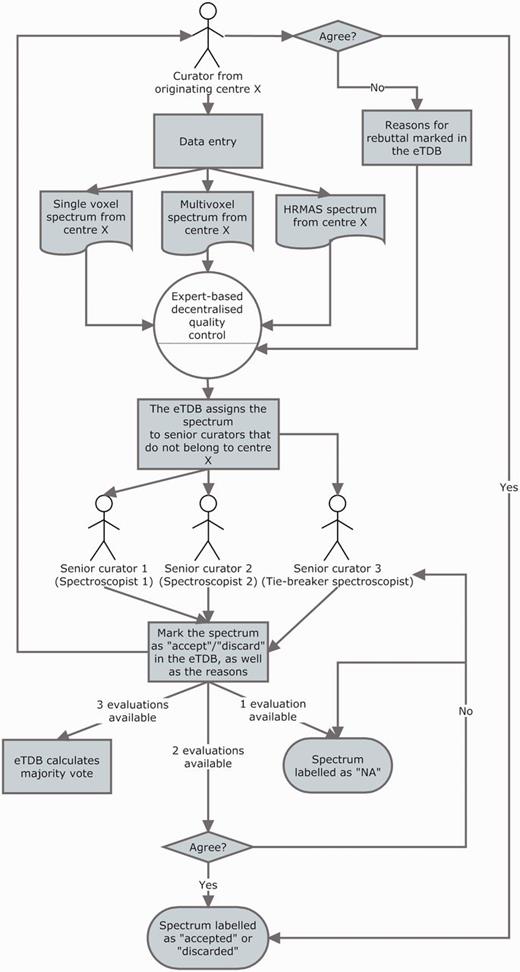
The three-expert system for spectroscopic QC of SV, MV and HRMAS. Two ES decided on the quality of the spectrum based on its empirical examination and a third one, a potential tie-breaker evaluated in case of disagreement. The spectrum could be accepted or discarded, and the instrumental reasons for it could also be stated if deemed necessary, e.g. poor signal-to-noise ratio (SNR), insufficient water suppression or signals from the scalp lipids (12). The system calculated, which was the majority vote and filled in the consensus decision that constituted another searchable meta-data field. It was as well possible that the original spectroscopist rebutted the final consensus decision and then, each of the ES could also change their verdict and state why they had changed their opinion. All these actions had specific meta-data fields. For SV spectra, the fields for ES validation were replicated, because depending on the format, it was possible to have an automatically processed spectrum or not, in which case, a manually processed one had to be entered. For MV data, because there was no automatic processing pipeline available, only expert-based meta-data fields were available. HRMAS validation was performed similarly to MV.
The clinical data validation (CDV) was entirely expert-based and performed at a central review facility in Nijmegen, The Netherlands (Figure 6 and Supplementary Figure S4). It involved not only experts from different medical disciplines (neurosurgeons, radiologists and neurooncologists) to review the MRI and clinical information for each case, but also a neuropathologists’ panel, to ensure first, diagnostic certainty (13–16) and second, that the voxel of the SV and MV study had been positioned according to protocols, covering the most malignant part of the tumour, avoiding clear necrosis, oedema or cystic areas (17), and co-localized with the area from which the subsequent biopsy had been obtained.
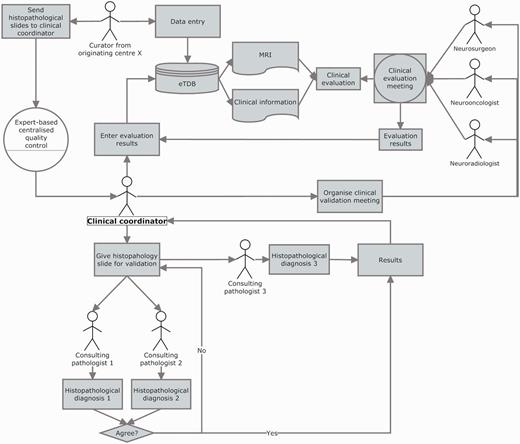
Clinical data validation. The review was internally coordinated by a clinical coordinator that collected all the results and entered them into the eTDB. This person was in charge of gathering the results of the clinical meetings, organizing the slides distribution among neuropathologists, collecting their diagnoses and entering all this meta-data for all the centres into the eTDB. In this case, the permission schema for this user was that of a senior curator and specifically tailored for this sole purpose.
In both situations (ES and clinical coordinator), the system checked the user’s profile against a table of permissions stored in the database, setting to visible/not visible or editable/non-editable, the corresponding edit or query fields on the interface. A set of complex queries, based on the permissions enabled to the user, retrieved all the experiments/cases pending evaluation and presented the expert with a list of the pending experiments awaiting evaluation.
The microarray QC was solely based on the text parameters associated to five parameters extracted from RPT (report) files. In this case, a decision rule was implemented that automatically checked these values for deciding which microarrays passed the QC and which one did not.
Summary results of the QC are displayed in Figure 7.
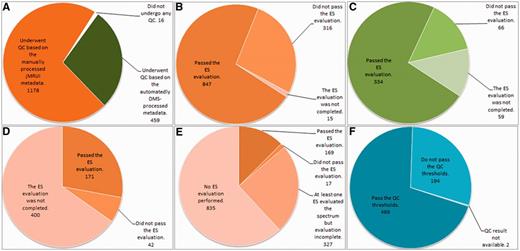
Summary results of the QC over the different data types. (A) Number of cases undergoing SV QC; (B) Results of the SV QC over the spectra that had been manually processed with jMRUI. (C) Results of the SV QC over the spectra that had been automatically processed with the DMS. (D) Results of the MV QC, all processing being manual. (E) Results of the HRMAS QC, all processing being manual. (F) Results of the automated microarray QC.
With respect to quality assessment (Figure 4), a protocol was set up, essentially as in previous study (7). The data manager engaged partners in a quality assessment (QA) round through the main eT e-mail list and set a deadline for sending out a summary of the results found. Each case could be labelled in each of the data types sections, indicating that it had undergone the QA as ‘the section is complete’ (Supplementary Figure S5). Detailed results of each QA round were kept off-line for the records and reports were written by the data manager and circulated among partners.
Defining metrics to quantify database contents
Monitoring how the database was being populated was a key issue, with respect to the specific coordination of the QC and QA activities, project coordination, and the periodic reports sent to the EU.
Being able to answer the simple question, ‘How many cases do we have in the database?’ was not a simple task itself, which could be further complicated to the ‘How many usable cases do we have in the database?’ which was periodically asked by the responsible of data analysis and by the coordination team of the project.
Why should this be a difficult-to-answer question? Or, what is ‘a usable case’ in fact? A case should—theoretically—have all the different data types set up by the acquisition protocols (clinical, SV, MV, HRMAS and microarray data), but the reality ranged from the most extreme situation in which a user might create a case and never upload any other information again, to varying degrees of completeness. For example, certain data types (e.g. only microarrays) might be entered by a specific centre for another centre or partner, because a certain platform (e.g. Affymetrix) might be available at a certain location or country only (the project started in 2004). Another example could be that partial datasets of one data type [e.g. SV spectra acquired only at one condition–echo time (TE)—of the two required by the acquisition protocols] because patients could not endure the whole set of scans (i.e. children). Other situations could be cases that were reasonably complete but that had not yet been validated by the clinical data validation (CDV) team and therefore lacked consensus diagnosis. Finally, cases could be found at the end of the project that had undergone QC but might not fulfil the requirements for one or more data types, to mention just a few examples.
Most importantly for the project’s development, the complexity of the information of each case, involved curators with expertise in in vivo MRS, neuropathology and neuroradiology. Therefore, completing the information for each case was essentially an asynchronous and team-based process. For example, a spectroscopist working in a clinical centre specialized in imaging would create a case in the eTDB, after patient ‘eTnnnn’ had undergone the MR exam, and the spectroscopist had checked that patient ‘eTnnnn’ fulfilled the inclusion criteria. Some days or weeks later, patient ‘eTnnnn’ might be operated in a different hospital in the same geographical area. Personnel at the University Department in the same partner would periodically collect all biopsy pieces stored in a liquid N2 container at the neurosurgery ward of this hospital. After that, a batch of biopsies would be processed at that University, and among those, the biopsy from patient ‘eTnnnn’ would be split in two if the piece was big enough for microarrays and HRMAS. Different departments (and in some cases, even different partners, depending on the availability of Affymetrix platforms with which the microarrays were processed) would perform the microarrays and HRMAS and enter the data into the eTDB once the RNA had been extracted and the microarray experiment performed. Sometime later, the whole MRI study would be gathered at the MR centre and the hospital and unstained biopsy slides would be collected from the pathology department, and sent to the central pathology review for validation. Clinical information would be followed up and the eTDB updated during the whole project for each patient with Karnofsky performance scores or survival data. This process would require the collaboration of a total of four or five different curators, apart from the clinicians, nurses and lab technicians involved in the study, but not directly interacting with the eTDB, having the curators edit permissions for the cases of the corresponding centre/s. Given this degree of complexity, it was deemed necessary to devise a set of simple metrics. These follow in the next section.
The indispensable dataset for quantifying usable data
The indispensable dataset (ID) (Figure 8) for each case was defined as the minimum amount of information that allowed one case to be usable for the project. This would happen when the case accomplished on one hand, the validation requirements set up in the QC protocols and on the other, when it could be used at least in one pattern recognition study, targeting in vivo MRS data-based classification studies. Fulfilling the ID did not exempt partners from the obligation to gather as much information as they could obtain for each case. However, eT aimed for the ideal situation in which all the information that could be uploaded in the eTDB was available (ideal dataset) as depicted in Figure 8.
![Indispensable (top) and ideal (bottom) datasets. For a case to fulfil the indispensable dataset, sufficient clinical information for performing the CDV had to be available. Sufficient clinical information was defined as the availability of: date of birth, sex, date of spectroscopy, at least one radiological classification (‘radiological diagnosis’ in the database), date of biopsy, paraffin section WHO (World Health Organization) classification (18, 19) (originating pathologist), relevant radiodiagnostic images [a representative T1-weighted image without contrast, a representative T2-weighted image and a representative image with voxel position(s) for the SV or the MV MRS]. In addition, the following data types were also needed to complete the indispensable dataset: one pair of MRS SV at short and long TE (except for rare cases such as childhood tumours and adults with small tumours where one echo time was acceptable), or one MRS MV at short TE, or one MRS MV at long TE, or HRMAS data, or one valid microarray.](https://oupdevcdn.silverchair-staging.com/oup/backfile/Content_public/Journal/database/2012/10.1093_database_bas035/1/m_bas035f8p.jpeg?Expires=1772760404&Signature=XzH9BP9w9cIuDWTA2Xt-lHjh1rgelZRcqZwh0H4USCn-GSZzXsNcbCFMSdP-AAZgQ1e2qxSAWXMGDYRDrvGYXGfdWXoI7jXOMiplKJ~fcxe0tgV2YA8bEoQrwjPQCTqpxu-qjmxX2kaMaPvG5~1YAD5zqg~DQBqL25IUeHxGE-HVzTEN~sUo5sX5BpvmJXx~y9wLEIrszWtle8EZWndUN1WPyWiCIWlESbsuW9en22hJ0RrJxzPRm7Flwu1OMXa5QNIe5DljkshgDseZbijD6SmZoen8m2KpBA36gNr62SOsZraOIkdE-CYHB6EgqfqD5HZKrjvryvEYDZuiTHIzBg__&Key-Pair-Id=APKAIYYTVHKX7JZB5EAA)
Indispensable (top) and ideal (bottom) datasets. For a case to fulfil the indispensable dataset, sufficient clinical information for performing the CDV had to be available. Sufficient clinical information was defined as the availability of: date of birth, sex, date of spectroscopy, at least one radiological classification (‘radiological diagnosis’ in the database), date of biopsy, paraffin section WHO (World Health Organization) classification (18, 19) (originating pathologist), relevant radiodiagnostic images [a representative T1-weighted image without contrast, a representative T2-weighted image and a representative image with voxel position(s) for the SV or the MV MRS]. In addition, the following data types were also needed to complete the indispensable dataset: one pair of MRS SV at short and long TE (except for rare cases such as childhood tumours and adults with small tumours where one echo time was acceptable), or one MRS MV at short TE, or one MRS MV at long TE, or HRMAS data, or one valid microarray.
On the other hand, it was also necessary to quantify the data entered in the eTDB with respect to the data types, and how these different data types were related to each other. In short, it was necessary to monitor whether a partner was uploading a more or less balanced set of data types for all their cases, or if there was any consistent lack of certain data types. The formulae in Table 2 account for this. Note that all indices were defined with respect to the number of cases with clinical information (NCCI). This happened because when a case was created, it was automatically searchable by clinical information even if no other information was filled in. Therefore, the maximum number of cases in the database was the NCCI. The NCCI is 1621, of which 304 had been migrated from the INTERPRET validated database (7) (http://gabrmn.uab.es/interpretvalidateddb/) through agreement between the two consortia. Therefore, 1317 cases were created in the eTDB as a result of the eT project.
| INDEX . | FORMULA . |
|---|---|
| Completeness | (NC data type × 100)/(NCCI) |
| Balancedness | {[(NCSV) + (NCMV) + (NCHRMAS) + (NCMA) + (NCMRI)] × 100}/(NCCI) |
| Complete pairedness | {[(NCSV) AND (NCMV) AND (NCHRMAS) AND (NCMA) AND (NCMRI)] × 100}/(NCCI) |
| In vivo MRS pairedness | [(NCSV) AND (NCMV) × 100]/(NCCI) |
| Ex vivo pairedness | [(NCHRMAS) AND (NCMA) × 100]/(NCCI) |
| SV and HRMAS pairedness | [(NCSV) AND (NCHRMAS) × 100]/(NCCI) |
| SV and microarrays pairedness | [(NCSV) AND (NCMA) × 100]/(NCCI) |
| MV and HRMAS pairedness | [(NCMV) AND (NCHRMAS) × 100]/(NCCI) |
| MV and microarrays pairedness | [(NCMV) AND (NCMA) × 100]/(NCCI) |
| INDEX . | FORMULA . |
|---|---|
| Completeness | (NC data type × 100)/(NCCI) |
| Balancedness | {[(NCSV) + (NCMV) + (NCHRMAS) + (NCMA) + (NCMRI)] × 100}/(NCCI) |
| Complete pairedness | {[(NCSV) AND (NCMV) AND (NCHRMAS) AND (NCMA) AND (NCMRI)] × 100}/(NCCI) |
| In vivo MRS pairedness | [(NCSV) AND (NCMV) × 100]/(NCCI) |
| Ex vivo pairedness | [(NCHRMAS) AND (NCMA) × 100]/(NCCI) |
| SV and HRMAS pairedness | [(NCSV) AND (NCHRMAS) × 100]/(NCCI) |
| SV and microarrays pairedness | [(NCSV) AND (NCMA) × 100]/(NCCI) |
| MV and HRMAS pairedness | [(NCMV) AND (NCHRMAS) × 100]/(NCCI) |
| MV and microarrays pairedness | [(NCMV) AND (NCMA) × 100]/(NCCI) |
The subindex indicates the data type. AND is used here as Boolean operator.
NC, number of cases; NCsv, number of cases with SV data; NCMV, number of cases with MV data; NCHRMAS, number of cases with HRMAS data; NCMA, number of cases with microarrays data.
| INDEX . | FORMULA . |
|---|---|
| Completeness | (NC data type × 100)/(NCCI) |
| Balancedness | {[(NCSV) + (NCMV) + (NCHRMAS) + (NCMA) + (NCMRI)] × 100}/(NCCI) |
| Complete pairedness | {[(NCSV) AND (NCMV) AND (NCHRMAS) AND (NCMA) AND (NCMRI)] × 100}/(NCCI) |
| In vivo MRS pairedness | [(NCSV) AND (NCMV) × 100]/(NCCI) |
| Ex vivo pairedness | [(NCHRMAS) AND (NCMA) × 100]/(NCCI) |
| SV and HRMAS pairedness | [(NCSV) AND (NCHRMAS) × 100]/(NCCI) |
| SV and microarrays pairedness | [(NCSV) AND (NCMA) × 100]/(NCCI) |
| MV and HRMAS pairedness | [(NCMV) AND (NCHRMAS) × 100]/(NCCI) |
| MV and microarrays pairedness | [(NCMV) AND (NCMA) × 100]/(NCCI) |
| INDEX . | FORMULA . |
|---|---|
| Completeness | (NC data type × 100)/(NCCI) |
| Balancedness | {[(NCSV) + (NCMV) + (NCHRMAS) + (NCMA) + (NCMRI)] × 100}/(NCCI) |
| Complete pairedness | {[(NCSV) AND (NCMV) AND (NCHRMAS) AND (NCMA) AND (NCMRI)] × 100}/(NCCI) |
| In vivo MRS pairedness | [(NCSV) AND (NCMV) × 100]/(NCCI) |
| Ex vivo pairedness | [(NCHRMAS) AND (NCMA) × 100]/(NCCI) |
| SV and HRMAS pairedness | [(NCSV) AND (NCHRMAS) × 100]/(NCCI) |
| SV and microarrays pairedness | [(NCSV) AND (NCMA) × 100]/(NCCI) |
| MV and HRMAS pairedness | [(NCMV) AND (NCHRMAS) × 100]/(NCCI) |
| MV and microarrays pairedness | [(NCMV) AND (NCMA) × 100]/(NCCI) |
The subindex indicates the data type. AND is used here as Boolean operator.
NC, number of cases; NCsv, number of cases with SV data; NCMV, number of cases with MV data; NCHRMAS, number of cases with HRMAS data; NCMA, number of cases with microarrays data.
Completeness accounted for the degree to which the different data types populated the database (Table 2). Balancedness is an indicative mean value for completeness of the different types and should be interpreted in view of the completeness values (Table 2). Complete pairedness is similar to the definition of ‘ideal dataset’ but it is less stringent because it only requires the presence of different data types and does not require availability of sufficient clinical information to perform the CDV. Partial pairedness measures (in vivo, ex vivo, etc.) indicate the percentage of cases for which two or more data types can be found. Results of the application of the main indices are shown through Venn diagrams in Figure 9. It is worth noting here that as INTERPRET did not gather HRMAS or microarrays, by default the corresponding 304 cases did not fulfil the complete pairedness standards of eT and the maximum achievable complete pairedness was 81% (1321 of 1621 cases). The explanation for such a low complete pairedness was the difficultness in gathering all the data types for each patient. The cases fulfilling this condition came exclusively from one participating centre, and correspond to the 15.2% of their cases (26 out of 171).
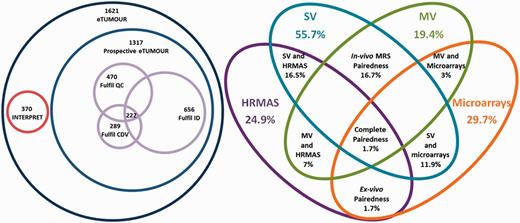
Venn diagrams of the main metrics defined. Left: number of cases by origin (INTERPRET or prospective eT), with numbers fulfilling the QC, the CDV and the ID and the intersection of all three. Size of Venn diagrams is proportional to the number of cases. Right: different pairedness values (intersections), with completeness for each data type. Values are expressed in percentage of cases from the total 1621. Size of Venn diagrams is not proportional to percentages.
Discussion
The eT project developed and populated a database, the eTDB, with multi-centre, multi-dimensional data from 1621 brain tumour patients, with a variable degree of completeness and quality.
The most important question faced in the project, from the data management point of view was, ‘how many cases are there in the eTDB?’. A synthetic and potentially unsatisfactory answer would be: ‘it depends… on which type of user you are and what you are looking for’. Is the potential user looking for cases that someone can use? Or for cases with all possible data types, a specific data type, necessarily of good quality and a consensus clinical diagnosis? Is the user familiar with the data types and prefers to make his/her own selection or is the user a mathematician looking for a carefully trimmed-off dataset ready for feeding into the latest machine learning algorithm? Does the user want to go translational and is looking for a collection with multi-dimensional ex vivo/in vivo data? Does the user need to download ‘everything’?
Setting up a metrics battery served the purpose of quantifying its contents, monitoring the data entry and curation processes. It might be argued that the application of certain metrics as inclusion criteria, such as, e.g. the need for CDV or the availability of the ID, caused the drop in the cases included with respect to the theoretical maximum of 1621 to 222, by joint probability (several events having to occur at the same time) and the existence of bottlenecks (one or several of these events having a low probability). This is a frequent and down-played situation encountered in almost any project dealing with data: subsequent application of inclusion criteria causes a progressive reduction in the number of cases available, the only difference in eT being that all potential cases were entered into the database by default. Similar projects involving the creation of a database have also encountered this: just to mention a couple of examples, the pilot TCGA research network (20) gathered in 2008, 587 cases, of which only 91 (15%) passed just four QC criteria, i.e. >80% tumour nuclei in specimens, <50% necrosis, a secondary pathology assessment and DNA or RNA of enough quality. Application of a much larger set of inclusion criteria on a more diverse dataset left a similar number (222 cases or 13.7%). In the antecedent of eT, INTERPRET, a dataset containing only in vivo MRS (mostly SV) and clinical data, application of inclusion criteria also left 30% of the original cases in the ‘INTERPRET validated database’ (7).
A validated database has been shown to be extremely useful, e.g. to test new mathematical algorithms and the INTERPRET database with its 304 cases provides an example of this (9, 21–30). As artificial intelligence experts do not necessarily have the expertise to interpret most of the information available from a specialized database, a validated dataset (not even a database) can simplify the task of evaluating new algorithms. On the other hand, application of a different metrics collection might result in a different subset of cases that might be useful in future and presently unplanned research questions on the same patient population.
With respect to bottlenecks identified with the metrics, the most important was fulfilment of the ID; and from the QC point of view, completing the CDV evaluation. Despite the large amount of spectra judged individually by two or even three different persons, the decentralized system worked well, because most spectra passed QC (Figure 7). The only problem was when one ES failed to perform the evaluation of a batch of data (i.e. HRMAS), then that whole block would remain with incomplete QC, although the individual evaluations were still recorded and queryable. Incomplete QC was an issue in the centralized protocol, in which disagreement among pathologists could exclude a case from entering the list of those with ID, no matter how good and/or complete the rest of data associated to the case are. A similar situation was encountered in the TCGA glioblastoma pilot (20). It has to be stressed though, in the context of brain tumours, and more specifically, of gliomas, this is a widely acknowledged fact (13–16, 31, 32). Therefore, the take-home message would be to apply a QC that is as de-centralized and as automated as possible. Even so, unavoidable bottlenecks might appear, specific of the area involved (i.e. histopathological characterization of gliomas), and these should be identified and minimized as soon as possible with the help of metrics.
From the technical point of view, a lesson learned from the experience with the eTDB would be the importance of having proper automated processing and visualization tools available, especially now that molecular imaging techniques are becoming widespread (33). In this respect, data standards for properly describing the data, such as the DICOM structured reporting (http://www.pixelmed.com/srbook.html), HL7 (http://www.hl7.org/), DICOM for MRS (http://www.dclunie.com/dicom-status/status.html), minimum information about a microarray experiment (MIAME) (34, 35) or minimum information for biological and biomedical investigations (MIBBI) (36), should provide an adequate starting point for future design of translational databases. However, in this project, an important part of data pre-processing and meta-data extraction had to be accomplished by reverse engineering, which indeed is not ideal. In this sense, a step forward, would be to adopt ontologies for the standardization of data representation and communication among different sources, enabling an easy integration to other systems. This option has been already explored in the area of brain tumour MRS (37), but not in the context of eT and its specific needs.
The eT project finished in August 2009 and as of 2012, all personnel specifically hired or involved in the project are working elsewhere, or funded to do other research. The database is still maintained ‘gratis et amore’ by a small team at the Universitat Autònoma de Barcelona (Spain) and a copy is saved at a server of the Universidad de Valencia (Spain), both available to those project participants that acquired the data, each centre being able to download their own data. Access to other partners is granted upon negotiation with the uploader partner. Access to external research groups for downloading the data was set to be carried out through the project management committee (through its former coordinator, Professor Celda from Universidad de Valencia) and the involved partners that gathered and curated these data. Access with view-only permissions (http://solaria.uab.es/eTumour/) is granted after request for a password, to the project management committee as well.
The accessibility solution chosen, despite not standard in the biological database area, is used in similar consortia, such as the dbGAP consortium (38) (http://www.ncbi.nlm.nih.gov/gap), where data access is carried out through a data access committee. It is also similar to the strategy adopted in INTERPRET (7), where the database has been already made accessible to more than 50 research groups to date, with view permissions and to three research groups with download permission upon request to the INTERPRET partners, with no applications having been turned down to date. The only requests to the groups with download permissions have been: use of data for the purpose stated, no unauthorized transfer to third parties and acknowledgement to data providers, similarly to dbGAP. Given the positive experience with the INTERPRET database and the lack of specific funding, a similar solution to INTERPRET could be adopted for the eTDB in the future.
All data in the eTDB are anonymized or de-identified, in accordance to the European Union 95/46/EC directive on data protection (http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=CELEX:31995L0046:EN:NOT) and national regulations of the involved countries, e.g. the Spanish Ley de Protección de Datos (https://www.agpd.es/), differently for example to other projects such as the TCGA, where the approach has been to set up two data tiers, one public with anonymous data and the second tier with non-de-identified data, subject as well to data transfer agreement.
Conclusion
The eT project gathered a complex dataset of in vivo and ex vivo data from brain tumour patients, in which QC checks at many levels were performed to ensure that subsequent statistical analysis was reliable. These checks were either automated (microarrays) or expert-based (spectroscopy and clinical information). For the latter, development of embedded data visualization tools was necessary. The decentralized system was less prone to generating bottlenecks limiting the number of data available. The development of a set of metrics was key to the proper monitoring of the eTDB contents.
Funding
The European Union (LSHC-CT-2004-503094). Partial support also from CIBER-BBN, which is an initiative funded by the VI National R&D&i Plan 2008–11, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund and Ministerio de Ciencia e Innovación in Spain (PHENOIMA: SAF2008-03323).
Conflict of interest. None declared.
Acknowledgements
Data gathering and curation is acknowledged for the following partners and persons: Universitat Autònoma de Barcelona, Cerdanyola del Vallès, Spain: Jaume Capellades, Jesús Pujol, Miriam Camisón, Teresa Delgado, Juana Martín, Milena Acosta, Indira Coronel, Daniel Valverde, Xavier Castells, Joaquin Ariño; Hospital Sant Joan de Déu, Barcelona, Spain: Antoni Capdevila; Radboud University, Nijmegen, The Netherlands: Arend Heerschap, Sandra Sprenger, Jack vanAsten, Jannie Wijnen, Pieter Wesseling, Albert Idema; St. George’s University of London, London, UK: Franklyn Howe, Alan James Wright, Kirstie Opstad, Greg Fellows; Medical University of Lodz, Lodz, Poland: Witold Gajewicz, Jan Fortuniak; Fundación para la Lucha contra las Enfermedades Neurológicas de la Infancia, Buenos Aires, Argentina: Jorge Calvar, Horacio Martinetto, Lina Núñez; Deutsche Krebsforschungzentrum, Heidelberg, Germany: Peter Bachert, Stefan Kirsch; Birmingham University, Birmingham, UK: Andrew Peet, Martin Wilson, Nigel Davies, Jane Crouch, Heather Crosby; The University of Nottingham, Nottingham, UK: Richard Grundy, Hazel Rogers; Cancer Research UK Cambridge Research Institute, Cambridge, UK: John Griffiths, Dominick McIntyre, Mary McLean, Madhu Basetti.
References
Author notes
†These authors contributed equally to this work.



