-
PDF
- Split View
-
Views
-
Cite
Cite
Stephanie M. Bunt, Gary B. Grumbling, Helen I. Field, Steven J. Marygold, Nicholas H. Brown, Gillian H. Millburn, the FlyBase Consortium, Directly e-mailing authors of newly published papers encourages community curation, Database, Volume 2012, 2012, bas024, https://doi.org/10.1093/database/bas024
Close - Share Icon Share
Abstract
Much of the data within Model Organism Databases (MODs) comes from manual curation of the primary research literature. Given limited funding and an increasing density of published material, a significant challenge facing all MODs is how to efficiently and effectively prioritize the most relevant research papers for detailed curation. Here, we report recent improvements to the triaging process used by FlyBase. We describe an automated method to directly e-mail corresponding authors of new papers, requesting that they list the genes studied and indicate (‘flag’) the types of data described in the paper using an online tool. Based on the author-assigned flags, papers are then prioritized for detailed curation and channelled to appropriate curator teams for full data extraction. The overall response rate has been 44% and the flagging of data types by authors is sufficiently accurate for effective prioritization of papers. In summary, we have established a sustainable community curation program, with the result that FlyBase curators now spend less time triaging and can devote more effort to the specialized task of detailed data extraction.
Database URL:http://flybase.org/
Introduction
One of the key sources of data within Model Organism Databases (MODs) is the primary research literature. The literature curators in a MOD extract biological data from research papers and convert it into a form suitable for loading into a database and display on a website. In many cases, the number of curators is not sufficient to curate all the papers relevant to their model organism, especially with the increasing number of papers published per year (Supplementary Figure S1) (1) and the advent of high-throughput studies. Therefore, it is essential to have a strategy to prioritize papers so that the fraction of papers that can be curated includes those with the data of most value to the community.
At FlyBase (http://flybase.org), the MOD for Drosophila genetic and genomic information (2), the data types that we prioritize include genetic data (e.g. new mutant alleles or transgenic constructs; phenotypic data; the first description of the function of a previously uncharacterized gene) and molecular data (e.g. new information about gene model structure; gene expression information) (Table 1). FlyBase curators have employed different strategies for prioritizing papers over the lifetime of the database. For the first 16 years, we stratified journals into priority groups based on a combination of the impact factor and prevalence of Drosophila genetic and genomic papers, and the majority of detailed literature curation was ordered using these groups. In 2008, we added a first-pass or ‘skim’ curation step into the prioritization pipeline, so that we now endeavour to quickly read all new Drosophila-related research papers to flag the occurrence of the data types prioritized by FlyBase. These flags can subsequently be used to prioritize papers for detailed data extraction (‘full’ curation). This new strategy has the advantage that a paper reporting valuable information on genes and reagents will be prioritized for full curation based on its significance to FlyBase and not on other less relevant factors, such as the journal it is published in.
| Data type flag . | Data presented in paper . |
|---|---|
| Drosophila reagents | |
| New allele or aberration | Generation of a new classical allele or chromosomal aberration in a Drosophilid genome. |
| New transgenea | Generation of a new transgenic construct. |
| Gene characterization | |
| Initial characterization | Initial characterization of a Drosophilid gene. |
| Merge of gene reports | Evidence suggesting the merge of two or more FlyBase Gene Reports. |
| Gene rename | New gene symbol or name for an existing gene in FlyBase. |
| Expression | |
| Expression in a wild-type background | New temporal or spatial expression data of any D. melanogaster gene in a wild-type background. |
| Expression in a mutant background | Expression data of any D. melanogaster gene in a mutant background or after environmental perturbation. |
| Phenotypes and interactions | |
| Phenotypic analysis | Novel phenotypic data. |
| Physical interaction | Physical interactions involving D. melanogaster proteins or nucleic acids. |
| Genome annotation data | |
| Changes to D. melanogaster gene model | New experimental data relevant to D. melanogaster gene model structure. |
| Changes to non-D. melanogaster gene model | New experimental data relevant to the gene model structure of non-D. melanogaster Drosophilid genes. |
| Mapping of features to genome | D. melanogaster molecular mapping data. |
| Cis-regulatory elements defined | Experimental definition of cis-regulatory elements of D. melanogaster genes. |
| Data type flag . | Data presented in paper . |
|---|---|
| Drosophila reagents | |
| New allele or aberration | Generation of a new classical allele or chromosomal aberration in a Drosophilid genome. |
| New transgenea | Generation of a new transgenic construct. |
| Gene characterization | |
| Initial characterization | Initial characterization of a Drosophilid gene. |
| Merge of gene reports | Evidence suggesting the merge of two or more FlyBase Gene Reports. |
| Gene rename | New gene symbol or name for an existing gene in FlyBase. |
| Expression | |
| Expression in a wild-type background | New temporal or spatial expression data of any D. melanogaster gene in a wild-type background. |
| Expression in a mutant background | Expression data of any D. melanogaster gene in a mutant background or after environmental perturbation. |
| Phenotypes and interactions | |
| Phenotypic analysis | Novel phenotypic data. |
| Physical interaction | Physical interactions involving D. melanogaster proteins or nucleic acids. |
| Genome annotation data | |
| Changes to D. melanogaster gene model | New experimental data relevant to D. melanogaster gene model structure. |
| Changes to non-D. melanogaster gene model | New experimental data relevant to the gene model structure of non-D. melanogaster Drosophilid genes. |
| Mapping of features to genome | D. melanogaster molecular mapping data. |
| Cis-regulatory elements defined | Experimental definition of cis-regulatory elements of D. melanogaster genes. |
The five categories used to group similar flags in the FTYP tool are shown in bold in the left-hand column above the individual flags in that group.
aThe ‘new transgene’ flag is not included in the analysis of the accuracy of community curated data as this flag was added to the author submission process after the set of 748 papers analysed had been submitted by authors.
| Data type flag . | Data presented in paper . |
|---|---|
| Drosophila reagents | |
| New allele or aberration | Generation of a new classical allele or chromosomal aberration in a Drosophilid genome. |
| New transgenea | Generation of a new transgenic construct. |
| Gene characterization | |
| Initial characterization | Initial characterization of a Drosophilid gene. |
| Merge of gene reports | Evidence suggesting the merge of two or more FlyBase Gene Reports. |
| Gene rename | New gene symbol or name for an existing gene in FlyBase. |
| Expression | |
| Expression in a wild-type background | New temporal or spatial expression data of any D. melanogaster gene in a wild-type background. |
| Expression in a mutant background | Expression data of any D. melanogaster gene in a mutant background or after environmental perturbation. |
| Phenotypes and interactions | |
| Phenotypic analysis | Novel phenotypic data. |
| Physical interaction | Physical interactions involving D. melanogaster proteins or nucleic acids. |
| Genome annotation data | |
| Changes to D. melanogaster gene model | New experimental data relevant to D. melanogaster gene model structure. |
| Changes to non-D. melanogaster gene model | New experimental data relevant to the gene model structure of non-D. melanogaster Drosophilid genes. |
| Mapping of features to genome | D. melanogaster molecular mapping data. |
| Cis-regulatory elements defined | Experimental definition of cis-regulatory elements of D. melanogaster genes. |
| Data type flag . | Data presented in paper . |
|---|---|
| Drosophila reagents | |
| New allele or aberration | Generation of a new classical allele or chromosomal aberration in a Drosophilid genome. |
| New transgenea | Generation of a new transgenic construct. |
| Gene characterization | |
| Initial characterization | Initial characterization of a Drosophilid gene. |
| Merge of gene reports | Evidence suggesting the merge of two or more FlyBase Gene Reports. |
| Gene rename | New gene symbol or name for an existing gene in FlyBase. |
| Expression | |
| Expression in a wild-type background | New temporal or spatial expression data of any D. melanogaster gene in a wild-type background. |
| Expression in a mutant background | Expression data of any D. melanogaster gene in a mutant background or after environmental perturbation. |
| Phenotypes and interactions | |
| Phenotypic analysis | Novel phenotypic data. |
| Physical interaction | Physical interactions involving D. melanogaster proteins or nucleic acids. |
| Genome annotation data | |
| Changes to D. melanogaster gene model | New experimental data relevant to D. melanogaster gene model structure. |
| Changes to non-D. melanogaster gene model | New experimental data relevant to the gene model structure of non-D. melanogaster Drosophilid genes. |
| Mapping of features to genome | D. melanogaster molecular mapping data. |
| Cis-regulatory elements defined | Experimental definition of cis-regulatory elements of D. melanogaster genes. |
The five categories used to group similar flags in the FTYP tool are shown in bold in the left-hand column above the individual flags in that group.
aThe ‘new transgene’ flag is not included in the analysis of the accuracy of community curated data as this flag was added to the author submission process after the set of 748 papers analysed had been submitted by authors.
Using skim curation as an initial triage process does however have disadvantages, primarily that it is time consuming (a curator typically averages two to three papers an hour) and often results in a duplication of effort, as each fully curated paper is read more than once. Over 2000 Drosophila-related papers are now typically published each year (Supplementary Figure S1) and there are many demands on curators in addition to curation of journal publications (e.g. responding to user requests; helping to enhance the design of the public website; developing methods to capture new data types of interest to the community). We would prefer to devote curation time to the specialized task of full curation rather than the initial triaging of papers and have thus been exploring alternative methods to prioritize them. Other MODs facing this problem have chosen to select articles from journals with the highest impact factor first (3), used automated methods such as text mining to flag data types and genes (4–6) and encouraged community-based annotation (3, 5).
Here, we describe how FlyBase is engaging authors to carry out skim curation of their recent publications. We began by setting up a simple web interface that allows authors to perform skim curation, indicating that doing so would speed up the incorporation of their data into FlyBase. The tool was announced on the FlyBase home page and promoted in FlyBase workshops, but relatively few authors used the tool. We then solicited author participation more directly, by e-mailing the corresponding author soon after publication and requesting that they use the web interface to skim curate their paper. Our e-mail message further simplifies use of the tool by linking to a form pre-populated with citation data, while its timing capitalizes on the enthusiasm authors feel when their papers first appear in press. This approach has been remarkably successful, resulting in 44% of new Drosophila-related papers being skim curated by authors. We describe the pipeline we devised to e-mail the authors and we assess the effectiveness of author curation for triaging papers.
Overview of the literature curation pipeline
Publications of all types that may contain Drosophila-related information are identified by a weekly semi-automated literature search of the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/) (Figure 1). The citation data for each publication verified to contain Drosophila-related information are then uploaded into the bibliography of the FlyBase database. Prior to integrating community curation into the pipeline, each new primary research paper was subsequently quickly read (‘skimmed’) by FlyBase curators (Figure 1a). Skim curation has two aims: first, to curate a limited amount of data (identifying the main genes studied in the paper and recording if an antibody has been generated), and secondly, to record the types of data contained within the publication using a defined set of ‘flags’ (Table 1). The skim-curated gene and antibody information is displayed on the FlyBase website at the next update, independent of any further data extraction. The data type flags are stored internally in the FlyBase database and are used by curators to prioritize data-rich papers for full curation.
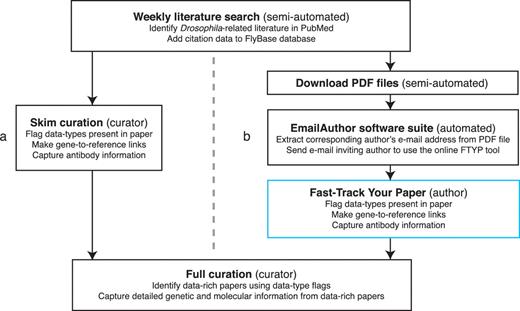
Literature curation pipeline before (a) and after (b) integrating community curation. A weekly search of the PubMed database identifies recent Drosophila-related publications. Newly identified papers subsequently undergo skim curation, which assigns data type flags and captures a limited subset of curated information (genes studied and antibodies generated). The data type flags are used to identify data-rich papers which are prioritized for full curation. The skim curation step previously carried out by FlyBase curators (a), has been replaced by community curation (b) by adapting the pipeline. First, we now download the PDF file of each new publication (currently possible for 89% of new papers). Secondly, we developed the EmailAuthor software suite, which is used to automatically e-mail the corresponding author of new papers. Finally, authors who have been e-mailed use the FTYP tool to skim curate their paper.
Although the majority of papers selected for full curation are identified through the skim curation pipeline, there are other circumstances that can result in a paper being prioritized for detailed data extraction. For example, curators may identify an unprioritized paper that describes the generation of a reagent when fully curating a prioritized paper or may be notified of a paper that describes a newly acquired stock by a Drosophila stock centre. Once a paper has been prioritized, the full curation process is the same: the curator captures all of the relevant data from the publication in a curation record, using a series of data-entry forms. After undergoing quality-control checks, these curation records are loaded into the FlyBase database at weekly intervals.
Design of a web-based community curation tool
The first step in enabling the research community to contribute to literature curation was the development of a web-based curation interface, which we have named ‘Fast-Track Your Paper’ (FTYP). The FTYP tool consists of six steps, which guide the user through the complete skim curation process (Figure 2; http://flybase.org/submission/publication/). Authors or other interested researchers can search for and confirm publication details, enter contact information (name, e-mail address and whether they are an author of the publication) and then skim curate the paper. Data submitted by authors and non-authors are treated identically.
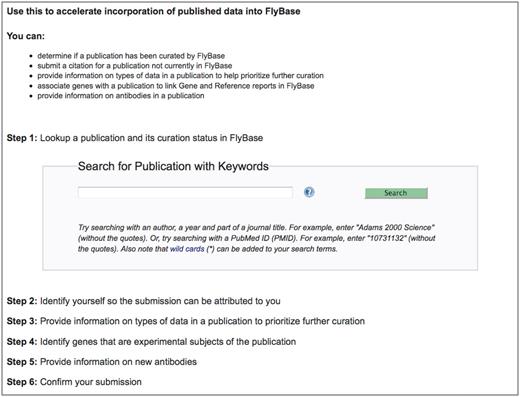
The Fast-Track Your Paper tool. The first page of the FTYP tool, listing the six steps that guide the user through the complete community curation process.
User-submitted data do not require manual inspection by a curator prior to loading into the FlyBase database. This is because we designed the FTYP tool to guide the user through the skim curation process as much as possible, rather than simply allowing them to enter their own choices in free text boxes. For example, the user chooses the appropriate data type flag(s) from a list rather than entering their own suggestions and enters gene symbols or names into a search field before selecting any appropriate matches from the resulting hit list. A user can enter the symbol of a gene unknown to FlyBase as free text if no matches are found. These ‘new’ genes are listed in an internal database field so that curators can determine whether the gene really is new to FlyBase at the time of full curation.
New user-submitted data are loaded into the FlyBase database weekly, in an identical format and process to data generated by FlyBase curators. Once loaded, user-submitted triage data are immediately available to curators to help prioritize papers for full curation, while the gene and antibody data become visible on the public website at the next update, 3–12 weeks after the original user submission (depending on when the data are submitted relative to the FlyBase release cycle).
The FTYP tool was launched on the FlyBase website on 26 February 2009. This first version of the tool was available via a link in the menu bar of each FlyBase page. The link directed the user to Step 1 of the form, where they could search for a publication and start the skim curation process. Over an 18-month period, 159 user submissions were received, 150 (94.3%) of which were from an author of the publication. This equates to ∼9 user submissions per month which was too few to have a significant impact on the literature curation pipeline. During this 18-month period, we tried to increase usage of the tool by adding a more visible link on the FlyBase home page and by having a commentary article highlighting the tool, but to no effect.
Development of software to automatically e-mail authors of Drosophila-related research papers
To try to increase the use of the FTYP tool, we decided to directly invite authors of recently published papers to complete a version of the form tailored to their publication. Figure 1b outlines the changes made to our literature curation pipeline to implement this new approach.
The first change was to add a step to our weekly PubMed literature search to download all new Drosophila-related publications in electronic (PDF) format from journals to which we have electronic access.
Secondly, we developed an automated software suite, named ‘EmailAuthor’, which generates and sends a set of personalized e-mails for any list of papers (Figure 3). For each paper on the list, EmailAuthor extracts the corresponding author’s e-mail address from the relevant PDF file (if it is available) and then uses the citation data for each paper (retrieved from the FlyBase database) to compose an e-mail message to the corresponding author. Each e-mail contains a personalized hyperlink to the FTYP tool, directing the author to a pre-filled form that displays their e-mail address and publication details, bypassing the previous requirement for authors to search for or fill in this information. Once the author has confirmed that this information is correct, they can progress directly to Step 3 of the tool and then complete the author submission as before. By integrating EmailAuthor directly after the weekly PubMed literature search, we contact authors as soon as possible after the publication of their paper.
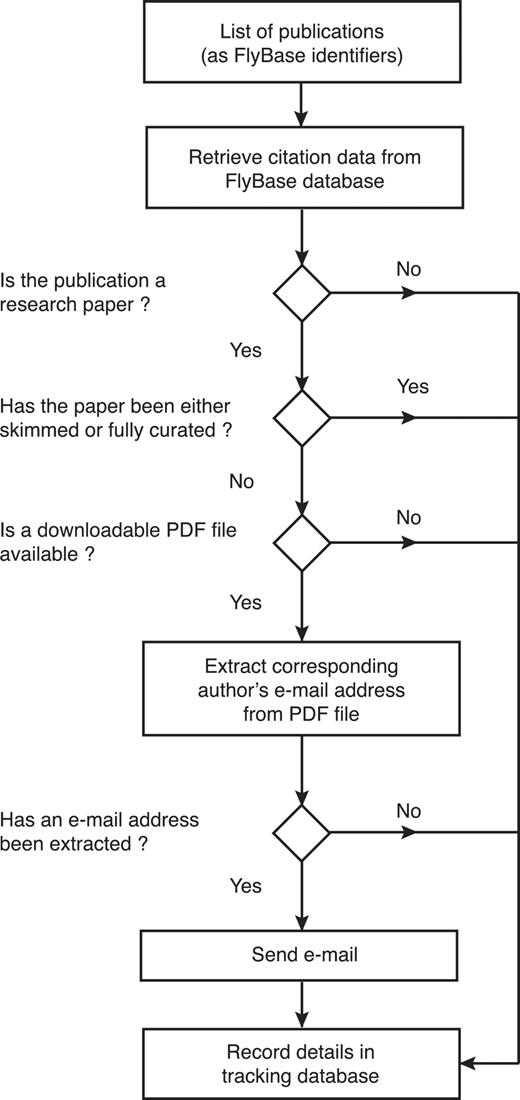
Workflow of the EmailAuthor software suite. For each publication, the software first checks its type and curation status using information stored in the FlyBase database. If it is a research paper that has not yet been triaged and a PDF file corresponding to the paper is available, the software attempts to extract the corresponding author’s e-mail address from the PDF file. If this is successful (97% of cases), an e-mail is sent to the extracted e-mail address. At each decision point, the information is stored in a tracking database.
To prevent duplication of effort, FlyBase curators stopped skimming newly published papers once we began e-mailing authors. We also added two checks to the pipeline to minimize overlap; for example, to prevent an author skim curating a recently published paper that has already been prioritized for full curation because it describes a stock that has been newly acquired by a stock centre. First, EmailAuthor only sends an e-mail for papers that have not already been skimmed or fully curated. Secondly, the FTYP tool now checks the curation status of the publication entered by the user; if the publication has already been fully curated or skimmed, the user is redirected to a page that thanks them and indicates the curation status of the paper, rather than allowing them to fill out the form.
Response rate to direct e-mailing
We have analysed the response rate for the first 9 months since we started our weekly direct e-mailing of authors on 18 October 2010. During this period, we sent 1282 e-mails to corresponding authors and received 568 responses via the personalized hyperlink. This equates to an overall response rate of 44.3% (Figure 4a), which is comparable to the rate of 40% obtained by WormBase using a similar approach (5). A further 14 author submissions were completed after we had sent an e-mail, but not through the personalized hyperlink. If these submissions were in response to the e-mail the response rate increases to 45%. Regardless, it is clear that our strategy of directly e-mailing authors increased the rate of community curation from ∼9 submissions per month to an average of ∼63 submissions per month.
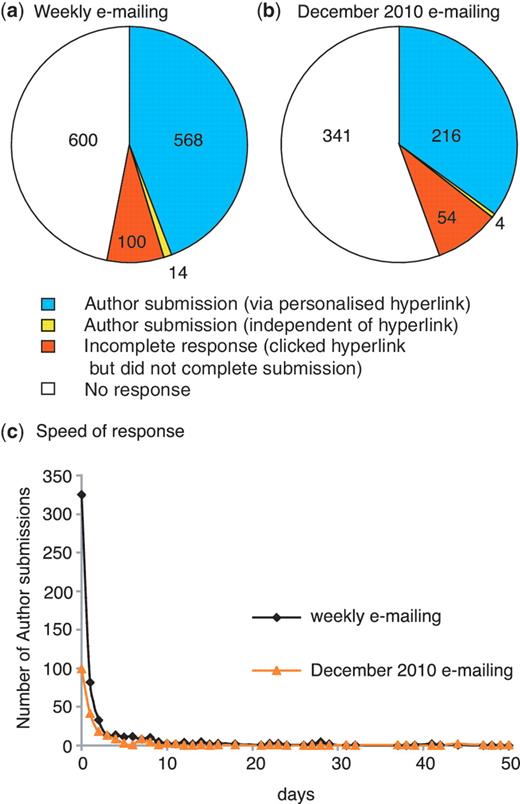
Author response to direct e-mailing. Overall response to (a) weekly e-mailing (corresponding author e-mailed <2 weeks after the entry for the published paper appeared in PubMed) and (b) single e-mailing to authors of untriaged papers carried out in December 2010 (in this case a PubMed entry for the published paper had existed for 2–13 months prior to e-mailing the corresponding author). The number of papers in each category is shown. (c) Speed of author response: number of days between author being sent e-mail and completing the author submission.
We were also able to test our hypothesis that authors of newly published papers would be more willing to assist with curation for FlyBase. In December 2010, we used the EmailAuthor program to contact corresponding authors for all untriaged papers published in 2010 prior to our starting the weekly direct e-mailing. For these papers, a PubMed entry for the published paper had existed for 2–13 months prior to e-mailing. The response rate via the hyperlink to this one-off e-mailing was lower than the average weekly e-mailing response, at 35.1% (Figure 4b). Thus, it appears that authors of recently published papers are most responsive to outreach.
Analysis of the delay between the author being sent an e-mail and then using the FTYP tool to complete a submission indicates that three-quarters of the authors who respond do so within 2 days of receiving the e-mail (Figure 4c) and that the speed of response to the weekly e-mails and to the one-off e-mailing in December 2010 follows the same pattern. For submissions completed via the personalized hyperlink in response to the weekly e-mailing, the corresponding author filled in the form themselves in 91% of cases, while 72.5% of the remaining responses were from the first author of the paper. The corresponding figures for the December 2010 e-mailing are 84% and 67.6%, respectively.
Non-solicited use of the FTYP tool in the 9 months following the start of weekly e-mailing resulted in 100 submissions (95% from authors of the publication). This represents an average of 11 voluntary submissions per month, a slight increase on the voluntary use of the tool before we started e-mailing (∼9 submissions per month). However, this increase can be entirely accounted for by 17 cases where an author has apparently checked the curation status of an earlier publication and completed a voluntary submission shortly after completing a separate author submission in response to being e-mailed about a different, recently published paper. Thus, the baseline voluntary use of the FTYP tool completely independent of an author receiving an e-mail does not appear to have increased since we started the weekly e-mailing process. This illustrates the importance of direct contact with authors in order to achieve a high level of community curation.
Accuracy of the community curated data
A system that relies on authors to carry out the initial skim curation of newly published papers is only effective if authors select the data type flags accurately and curate the gene and antibody data correctly. To assess the accuracy of the community curated data, FlyBase curators carried out detailed curation of 748 papers that had been community curated using the FTYP tool, regardless of whether authors assigned any data type flags. We corrected any errors in the author-submitted data during the full curation process, allowing us to assess the accuracy of the author-submitted data by comparing the original author submission record with the data currently held in FlyBase.
Authors triaged their paper completely accurately in 59.9% of cases, selecting all the correct flag(s) corresponding to the data types presented (316 papers) or alternatively not checking any flags because none of the data types currently flagged were presented (132 papers) (Figure 5a). Thus, ∼40% of papers are not triaged completely accurately by authors. However, this does not translate directly into the fraction of papers that would have incorrectly been present in or absent from the prioritized list for full curation if we relied solely on these author-assigned flags, because of the way we use the flags to prioritize papers. For genetic data, we prioritize papers based on the total number of relevant data type flags and thus inaccuracies will mostly affect the position of papers in this priority list, not whether or not they appear on the list at all: in the set of 748 papers analysed, only 0.5% (4 papers) would not have been prioritized if we had relied solely on the flags submitted by authors (i.e. false negatives) and only 9.9% (74 papers) would have been prioritized incorrectly (i.e. false positives).
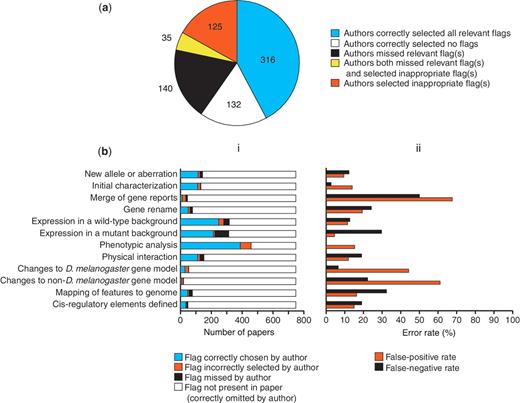
Accuracy of author-submitted data type flags. (a) Accuracy at the level of the whole paper. The number of papers in each category is shown. (b) Accuracy on a flag-by-flag basis. (i) Frequency of occurrence and accuracy of selection of each data type flag. (ii) Error rates for selection of each data type flag.
Figure 5b shows the accuracy of flags assigned by authors for each individual data type. There appears to be no correlation between authors missing a particular flag and instead choosing another flag. For most data types where the false-positive rate is high, that data type is rare, meaning that flags incorrectly selected by authors generally have little impact on the curation pipeline. However, the false positives for one data type, ‘Phenotypic analysis’, do have a relatively large effect on the prioritization pipeline, because it is a common data type that is present in just over 50% of papers. Thus, despite a relatively low false-positive rate for this data type (just over 15%), there were 50 papers where a false-positive ‘Phenotypic analysis’ flag was the only flag selected. We have been able to compensate for this by adjusting the prioritization of papers for full curation, so that those papers where the only genetic data type flag is ‘Phenotypic analysis’ are put at the bottom of the priority list, below all other papers with a different single genetic data type flag.
High false-negative rates are potentially more problematic than high false-positive rates, as they may result in data-rich papers being overlooked for full curation. In most cases, data types with higher false-negative rates are present less frequently in papers (Figure 5b), so relatively few data-rich papers will be missed. However, flagging of the ‘Expression in a mutant background’ data type shows a relatively high false-negative rate (almost 30%) and this data type is present in 40% of the 748 papers examined. Fortunately, this data type is often associated with the ‘Expression in a wild-type background’ data type (64.4% of cases in the set of 748 papers) and the false-negative rate for flagging this latter data type is lower (only 12.6%). Both types of expression curation are carried out by the same group of curators thus reducing the overall false-negative rate.
We found that 673 of the 748 author-submitted papers analysed contain gene information after full genetic data curation. Authors chose to fill in gene information for 69.8% (470/673) of these papers, resulting in a total of 4614 gene to paper links. Curators removed incorrect gene associations from only 4.1% (31/748) of the author-submitted papers during full curation. Gene data curation by authors is thus highly accurate and was carried out for over two-thirds of those papers where it was possible.
Authors added 93 antibody statements to the set of 748 papers. The false-positive rate was 16.1% (15/93 statements were removed during full curation). In 80% (12/15) of these cases, the authors had used an antibody to the gene product in the paper, but the antibody was not generated in that publication. In addition, the authors missed 31 antibody statements, a false-negative rate of 28.4%. In 80.6% (25/31) of these cases, the authors had not filled in any genes for the publication, so they would not have seen the subsequent question asking if the publication reported the generation of an antibody.
Conclusions and future plans
Through the development of a web-based curation tool and direct e-mailing of authors, we have achieved a high rate of community curation, with authors of recently published papers being most responsive to the e-mails (44% response). We also have evidence that directing authors to a particular publication (via a personalized hyperlink) increases the likelihood of a productive response (Figure 6); although a general e-mail (which did not direct authors to a particular publication) sent to the community shortly prior to commencing the weekly e-mailing resulted in increased usage of the FTYP tool, this was largely unproductive, consisting of cases where a user attempted to curate a paper that had already been fully curated or skimmed. Our findings thus fully support the proposal of Mazumder et al. (7), that to successfully engage the community in curation it is necessary to proactively solicit contributions and to provide clear instructions on what requires curation. Authors have provided highly accurate gene data for a large fraction of papers, resulting in valuable links between recently published papers and the main genes studied in them on the FlyBase website. Importantly, selection of data type flags by authors is sufficiently accurate to prioritize papers effectively for full curation: the number of false negatives and false positives is either low or can be compensated for by co-occurrence of some data types or by other aspects of the curation pipeline.
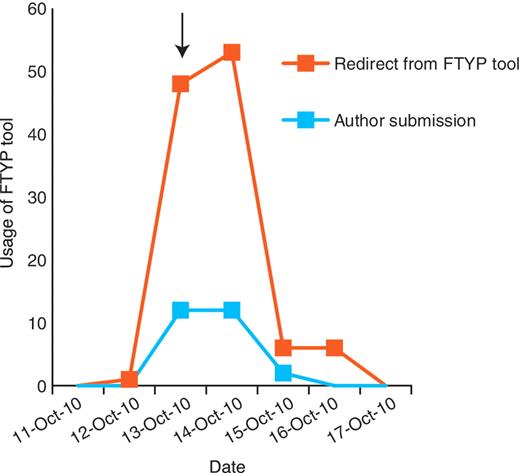
Community curation is most productive when authors are directed to a particular publication. A general e-mail was sent to the Drosophila research community on 13 October 2010 (arrow), alerting them that we would be starting the weekly direct e-mailing the following week. This resulted in a small increase in successful author submissions, but resulted in a larger increase in unproductive redirects from the FTYP tool, where authors attempted to curate a paper that had already been skimmed or fully curated and were redirected to a page thanking them for their effort.
We plan to expand the scope of community curation by also e-mailing the corresponding authors of newly published review articles as part of the weekly e-mailing process. For FlyBase, full curation of reviews consists of simply recording the gene(s) that are the subject of the review. Therefore, the process will be simpler for authors than skim curation of papers because there will be no need select triage flags. This community effort will be particularly valuable as due to limited resources, FlyBase curators no longer routinely curate reviews and the community will effectively be fully curating the review.
Community curation has already replaced the need for FlyBase curators to skim curate over 40% of newly published papers. This equates to ∼2–3 months of curator time per year, thus freeing up considerable time for full data extraction. However, at present FlyBase curators still skim curate those papers that are not curated by the authors: we currently skim curate these papers 8 weeks after the original e-mail was sent, to minimize the likelihood of any overlap with author curation. To further reduce the time that curators need to spend on skim curation, future objectives include increasing the fraction of newly published papers curated by the community, further increasing the accuracy of data type flagging by authors and devising a strategy to automatically triage those papers not curated by their corresponding author.
To try to further increase the fraction of papers that are community curated we have recently reworked the wording and organization of the e-mail that authors receive, clarifying the benefits of completing a submission. We now also send a second ‘reminder’ e-mail, 2 weeks after the original one, to those authors who have not completed a submission using the FTYP tool. The rapid response of the majority of authors who complete a submission after receiving the first e-mail (Figure 4c) suggests that this follow up e-mail may prove effective in increasing the number of author submissions and also means that we can easily assay whether or not this second e-mail is effective in increasing the overall response rate. We may also increase the response rate if we can improve the usability of the tool, particularly if we can minimize incomplete submissions (8% of cases for e-mailed authors, Figure 4a and b). To this end, we plan to analyse the session data stored by the FTYP tool for these partial submissions to try to determine which step(s) are causing particular problems so that we can then attempt to improve their usability. We also plan to improve the gene selection step of the FTYP tool so that authors of high-throughput papers can submit lists of genes that are studied in their paper using FlyBase identifiers instead of searching using gene symbols or names.
To try to improve the accuracy of flagging for those data types with the highest false-positive and false-negative rates (‘Merge of gene reports’ and changes to both D. melanogaster and non-D. melanogaster gene models) we plan to change the FTYP form so that the examples for these particular flags are shown by default (currently a user must click on the help button next to the flag to see these). We can then monitor whether this results in an increase in the accuracy of flagging for these data types.
We are currently also investigating text mining methods (8) as a parallel approach to triaging papers for full curation, primarily for those papers where authors do not respond to our e-mail or where there is no e-mail address for the corresponding author (∼1.5% of papers). In the future, we envisage that a combinatorial approach will result in a high proportion of newly published papers being triaged for full curation without requiring the input of FlyBase curators, allowing us to devote more of our curation effort to detailed data extraction.
Funding
National Human Genome Research Institute at the U.S. National Institutes of Health [P41 HG000739]; Medical Research Council [G1000968]; Indiana Genomics Initiative. Hosting of the FlyBase website is supported in part by the National Science Foundation [OCI-1053575] through XSEDE resources provided by Indiana University. Funding for open access charge: National Human Genome Research Institute at the U.S. National Institutes of Health [P41 HG000739].
Conflict of interest. None declared.
Acknowledgements
The FlyBase Consortium comprises: William Gelbart, Nick Brown, Richard Cripps, Thomas Kaufman, Kathy Matthews, Maggie Werner-Washburne, Boris Adryan, Marta Costa, Lynn Crosby, Adam Dirkmaat, Gil dos Santos, David Emmert, Kathleen Falls, Helen Field, Josh Goodman, L. Sian Gramates, Gary Grumbling, Steven Marygold, Beverley Matthews, Peter McQuilton, Gillian Millburn, David Osumi-Sutherland, Harriett Platero, Laura Ponting, Susan Russo, Andy Schroeder, Ray Stefancsik, Susan St Pierre, Victor Strelets, Jim Thurmond, Susan Tweedie, J.D. Wong, Pinglei Zhou, Mark Zytkovicz. The authors would like to thank Paul Sternberg for encouraging us to pursue our idea of e-mailing authors of newly published papers to engage them in community curation. We would also like to thank members of FlyBase and three anonymous reviewers for critical comments on the manuscript, members of the FlyBase community curation subcommittee for helping to improve the design of the FTYP tool, and all the curators at the FlyBase-Cambridge site for helping to fully curate the set of 748 community curated papers so that we could analyse the accuracy of the author-submitted data. Finally, we would like to thank all the authors who have used the FTYP tool to complete an author submission and have made this community curation program a success.
References
Author notes
†These authors contributed equally to this work.
‡The members of the FlyBase Consortium are listed in the Acknowledgements.



