-
PDF
- Split View
-
Views
-
Cite
Cite
Akira R. Kinjo, Reiko Yamashita, Haruki Nakamura, PDBj Mine: design and implementation of relational database interface for Protein Data Bank Japan, Database, Volume 2010, 2010, baq021, https://doi.org/10.1093/database/baq021
Close - Share Icon Share
Abstract
This article is a tutorial for PDBj Mine, a new database and its interface for Protein Data Bank Japan (PDBj). In PDBj Mine, data are loaded from files in the PDBMLplus format (an extension of PDBML, PDB's canonical XML format, enriched with annotations), which are then served for the user of PDBj via the worldwide web (WWW). We describe the basic design of the relational database (RDB) and web interfaces of PDBj Mine. The contents of PDBMLplus files are first broken into XPath entities, and these paths and data are indexed in the way that reflects the hierarchical structure of the XML files. The data for each XPath type are saved into the corresponding relational table that is named as the XPath itself. The generation of table definitions from the PDBMLplus XML schema is fully automated. For efficient search, frequently queried terms are compiled into a brief summary table. Casual users can perform simple keyword search, and 'Advanced Search' which can specify various conditions on the entries. More experienced users can query the database using SQL statements which can be constructed in a uniform manner. Thus, PDBj Mine achieves a combination of the flexibility of XML documents and the robustness of the RDB.
Database URL: http://www.pdbj.org/
Introduction
All protein structural data must be deposited to the worldwide Protein Data Bank (wwPDB) (1) if they are to be published in scientific journals. After curation, the deposited data are saved in three different formats [PDB, mmCIF (2), and PDBML (3)] and are published at the FTP sites of the wwPDB members, which includes RCSB PDB (USA), PDBe (Europe) and PDBj (Japan) (together with BMRB [Biological Magnetic Resonance Bank]). Currently, there are over 66 000 entries available at the wwPDB. Although all the information about protein structures are available in mmCIF or PDBML files, the protein structural data are inherently complex, consisting of atomic coordinates, experimental methods and conditions, citation information and annotations of molecular entities to name a few, so that it is not always easy for the casual user to find necessary information. Therefore, each member of the wwPDB provides its own interface to the database via the worldwide web (WWW) (1).
As an enhancement to the PDBML format, we at PDBj have been developing PDBMLplus that is a super-set of PDBML and includes additional information related to each PDB entry such as manually curated experimental information and cross-reference to other databases. Based on PDBMLplus, we have recently developed PDBj Mine, a relational database (RDB) and its web interface (http://www.pdbj.org/; the list of web resources are provided in Table 1) to the PDBMLplus data. In this article, we describe the design and implementation of PDBj Mine. Although it is a RDB, PDBj Mine preserves the structure of PDBMLplus (an XML format) so that a user who is familiar with PDBML or PDBMLplus can easily construct SQL queries based on the hierarchical structure of XML. PDBj Mine is implemented using only free, open-source softwares so that interested power users can install the entire back-end system of PDBj Mine to make their own mirror sites of PDBj. The basic structure of PDBj Mine and the sample queries described in this article will help the user to construct more advanced queries against the PDBj to retrieve more useful information on protein structures.
| Resource . | URL . | Remarks . |
|---|---|---|
| wwPDB | http://www.wwpdb.org | |
| PDBj | http://www.pdbj.org | `Quick' search |
| PDBj Mine | http://service.pdbj.org/mine/ | SQL interface |
| Advanced Search | http://service.pdbj.org/mine/advanced.html | |
| PDBj Mine Help | http://doc.pdbj.org/help?PDBj%20Mine | Documentation |
| PDBMLplus | ftp://ftp.pdbj.org/XML/pdbmlplus | FTP site |
| PDBj Mine dump | ftp://ftp.pdbj.org/mine | FTP site |
| mmCIF dictionary | http://mmcif.pdb.org/ | |
| PDBML schema | http://pdbml.pdb.org/schema/pdbx-v32.xsd | |
| PDBMLplus schema | http://service.pdbj.org/mine/schema.html | |
| jV | http://www.pdbj.org/jv/ | Molecular graphics |
| JMol | http://www.jmol.org/ | Molecular graphics |
| PostgreSQL | http://www.postgresql.org | Version 8.4 series |
| Resource . | URL . | Remarks . |
|---|---|---|
| wwPDB | http://www.wwpdb.org | |
| PDBj | http://www.pdbj.org | `Quick' search |
| PDBj Mine | http://service.pdbj.org/mine/ | SQL interface |
| Advanced Search | http://service.pdbj.org/mine/advanced.html | |
| PDBj Mine Help | http://doc.pdbj.org/help?PDBj%20Mine | Documentation |
| PDBMLplus | ftp://ftp.pdbj.org/XML/pdbmlplus | FTP site |
| PDBj Mine dump | ftp://ftp.pdbj.org/mine | FTP site |
| mmCIF dictionary | http://mmcif.pdb.org/ | |
| PDBML schema | http://pdbml.pdb.org/schema/pdbx-v32.xsd | |
| PDBMLplus schema | http://service.pdbj.org/mine/schema.html | |
| jV | http://www.pdbj.org/jv/ | Molecular graphics |
| JMol | http://www.jmol.org/ | Molecular graphics |
| PostgreSQL | http://www.postgresql.org | Version 8.4 series |
| Resource . | URL . | Remarks . |
|---|---|---|
| wwPDB | http://www.wwpdb.org | |
| PDBj | http://www.pdbj.org | `Quick' search |
| PDBj Mine | http://service.pdbj.org/mine/ | SQL interface |
| Advanced Search | http://service.pdbj.org/mine/advanced.html | |
| PDBj Mine Help | http://doc.pdbj.org/help?PDBj%20Mine | Documentation |
| PDBMLplus | ftp://ftp.pdbj.org/XML/pdbmlplus | FTP site |
| PDBj Mine dump | ftp://ftp.pdbj.org/mine | FTP site |
| mmCIF dictionary | http://mmcif.pdb.org/ | |
| PDBML schema | http://pdbml.pdb.org/schema/pdbx-v32.xsd | |
| PDBMLplus schema | http://service.pdbj.org/mine/schema.html | |
| jV | http://www.pdbj.org/jv/ | Molecular graphics |
| JMol | http://www.jmol.org/ | Molecular graphics |
| PostgreSQL | http://www.postgresql.org | Version 8.4 series |
| Resource . | URL . | Remarks . |
|---|---|---|
| wwPDB | http://www.wwpdb.org | |
| PDBj | http://www.pdbj.org | `Quick' search |
| PDBj Mine | http://service.pdbj.org/mine/ | SQL interface |
| Advanced Search | http://service.pdbj.org/mine/advanced.html | |
| PDBj Mine Help | http://doc.pdbj.org/help?PDBj%20Mine | Documentation |
| PDBMLplus | ftp://ftp.pdbj.org/XML/pdbmlplus | FTP site |
| PDBj Mine dump | ftp://ftp.pdbj.org/mine | FTP site |
| mmCIF dictionary | http://mmcif.pdb.org/ | |
| PDBML schema | http://pdbml.pdb.org/schema/pdbx-v32.xsd | |
| PDBMLplus schema | http://service.pdbj.org/mine/schema.html | |
| jV | http://www.pdbj.org/jv/ | Molecular graphics |
| JMol | http://www.jmol.org/ | Molecular graphics |
| PostgreSQL | http://www.postgresql.org | Version 8.4 series |
Design
As mentioned above, we attempted in the design of PDBj Mine to embed the structure of PDBML into a set of relational tables as closely as possible so that the user who is familiar with PDBML can construct queries based on XPath expressions. Although the resulting design somewhat deviates from conventional RDB techniques and may sacrifice the efficiency of query processing, it makes possible to incorporate XML documents in a generic manner and, in practice, achieves reasonable efficiency.
We first review the PDBML and PDBMLplus formats, and then describe a method to represent the XML document structures using `pointers' to XML elements and attributes. Using the pointer representation, the contents of XML documents are stored in `XPath-based tables' as described below. After summarizing the general organization of the relational tables in PDBj Mine, we describe some practical tricks concerning keyword search and advanced searches.
Overview of PDBML and PDBMLplus formats
The PDBML format is the canonical XML format for PDB data (3), which is translated from the mmCIF format (2). Reflecting the structure of mmCIF, the structure of PDBML is relatively simple and of shallow hierarchy (Figure 1). In the mmCIF dictionary, related terms are grouped into categories (`xxxCategory' in Figure 1), and each category includes entities (`xxx' in Figure 1) specified by the items of that category (`yyy' and `zzz' in Figure 1). The attributes in each category element corresponds to the primary key of the element.
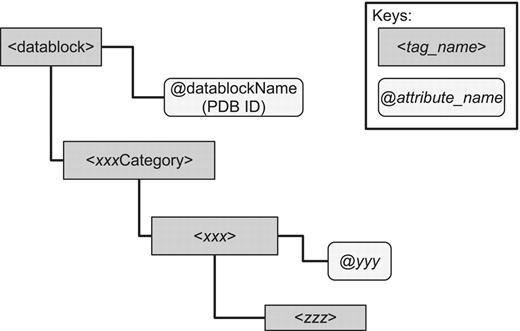
Basic structure of PDBML. Every PDBML file is enclosed in the `datablock' element with which the `datablockName' attribute indicating PDB ID is associated. Inside the datablock element, there are a number of elements corresponding to mmCIF category groups enclosed in the tags `xxxCategory' where xxx indicates one of the mmCIF category. There is only one xxxCategory element for each category group (`xxx' can be `entity', `citation', `struct' and so on). Inside an xxxCategory element, there are one or more elements enclosed in the xxx tag (this `xxx' is the same as in `xxxCategory' up in the hierarchy). For each xxx element, there are usually one or more attributes (denoted `@yyy' in the figure) which indicates the identifier of that element (e.g. `@id' in the entity, citation categories and `@entry_id' in the struct category). These attributes serve as the primary key for each category element. Under the xxx element, there are elements (denoted zzz in the figure) that are specific to the category (e.g. `pdbx_description' in the entity category, `pdbx_database_id_PubMed' in the citation category and `pdbx_descriptor' in the struct category). In the mmCIF dictionary, the attributes (such as `@yyy') and elements (such as `zzz') are referred to as items.
The PDBMLplus format is an extension of PDBML developed at PDBj (The XML schema is provided at http://service.pdbj.org/mine/schema.html). In PDBMLplus files, some manually curated experimental information as well as automatically associated cross-references to other database resources are added. Accordingly, there are a few additional elements that are not present in the canonical PDBML format. Some experimental information (e.g. mmCIF categories such as refine, reflns, software, citation, etc.) that are missing or incorrect in the original PDBML files are manually appended or corrected in PDBMLplus files by an annotator at PDBj who examines the original references (These fixes are reflected in the original PDB data in future releases or remediation). Other, mostly functional information cross-referenced by other databases to the PDB are automatically extracted every week. These include functional sites and gene ontology (http://www.geneontology.org) mapping obtained from the UniProt (4), catalytic site information from the Catalytic Site Atlas (5) and ligand binding sites identified in the PDB itself. Those annotations are appended in mmCIF categories (possibly extended) such as struct_site, struct_site_gen and struct_ref, or in a new category specific to PDBMLplus such as gene_ontology.
PDBMLplus files and its XML schema are available at ftp://ftp.pdbj.org/XML/pdbmlplus. The PDBMLplus file of each PDB entry can be also downloaded at the `Download/Display' page of the corresponding entry (Figure 3)
Representation of XML hierarchical structure of PDBMLplus
Each PDBMLplus file, being in an XML format, has a hierarchical tree structure defined by the PDBMLplus XML schema. In PDBj Mine, we model the RDB tables so as to preserve the XML structure of the PDBMLplus files. By doing so, we mimic a native XML database using an RDB so that the user of PDBML files can construct his query with less difficulty. To handle the contents of XML documents efficiently, PDBj Mine is designed to process XPath expressions in a straightforward manner. For each element or data in a PDBMLplus file, we assign a number (pointer) ordered in a depth-first manner along the tree structure (Figure 2). This method of assigning pointers to XML elements was originally proposed by Yoshikawa et al. (6) for their XRel method of storing and retrieving XML documents in a RDB. Unlike XRel, however, PDBj Mine stores each XML element type into a unique table named after its XPath expression. These tables are referred to as `XPath-based tables' in the following. For example, in Figure 2, the information of the datablock, entryCategory elements are stored in the tables named `E:/datablock' and `E:/datablock/entryCategory' (i.e. XPath name with the `E:' prefix), respectively. Attribute values and PCData (parsed character data) such as `1A00-noatom' in the `/datablock/@datablockName' attribute or `1' in the `/datablock/struct_asymCategory/struct_asym/entity_id' element are stored in the corresponding tables of the same name as the XPath name (without the `E:' prefix). Thus, there are two types of XPath-based tables: one for elements themselves, the other for attribute values and PCData. These two types of tables are referred to as `container table' and `content table', respectively, in the following. The basic structures of these tables are given in Table 2.
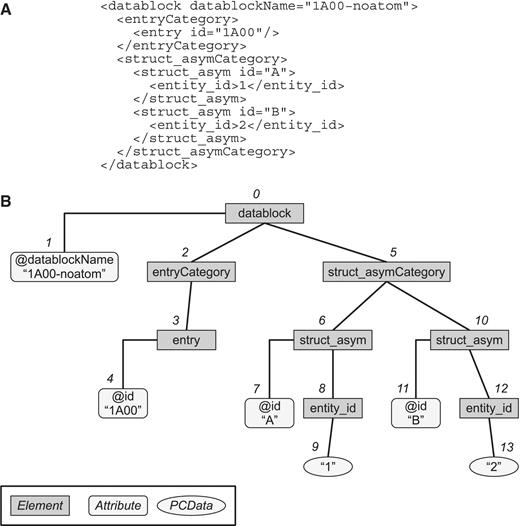
Document tree representation. (A) A sample PDBMLplus (XML) document. (B) The tree representation of the document in (A). Each element, attribute, or PCData is uniquely indexed with a pointer (from 0 to 13 in this example) according to the tree structure in the depth-first order.
| Column . | Type . | Example . |
|---|---|---|
| Container tables | `E:/datablock/entityCategory/entity' | |
| docid | INTEGER | 68 271 |
| pstart | INTEGER | 718 |
| pend | INTEGER | 737 |
| Content tables | `/datablock/entityCategory/entity/pdbx_ec' | |
| docid | INTEGER | 68 271 |
| pos | INTEGER | 727 |
| val | typeOfData | '1.1.3.9' |
| Xmldoc | xmldoc (this is the only instance) | |
| pdbid | TEXT | '1gof' |
| docid | INTEGER | 68 271 |
| doc | XML | '<datablock> … </datablock>' |
| kwd | TEXT | 'oxidation reduction … ' |
| mtime | TIMESTAMP | 2009-11-29 00:11:45+09 |
| Column . | Type . | Example . |
|---|---|---|
| Container tables | `E:/datablock/entityCategory/entity' | |
| docid | INTEGER | 68 271 |
| pstart | INTEGER | 718 |
| pend | INTEGER | 737 |
| Content tables | `/datablock/entityCategory/entity/pdbx_ec' | |
| docid | INTEGER | 68 271 |
| pos | INTEGER | 727 |
| val | typeOfData | '1.1.3.9' |
| Xmldoc | xmldoc (this is the only instance) | |
| pdbid | TEXT | '1gof' |
| docid | INTEGER | 68 271 |
| doc | XML | '<datablock> … </datablock>' |
| kwd | TEXT | 'oxidation reduction … ' |
| mtime | TIMESTAMP | 2009-11-29 00:11:45+09 |
There are two kinds of XPath-based tables for storing PDBMLplus data. The container tables contain the docid (PDB ID consisting of four alphanumerical characters interpreted as a radix 36 number), pstart (the pointer to the element) and pend (the maximum value of the pointers under that element). The table names of the container tables are XPaths prefixed with `E:'. The content tables contain docid, pos (the pointer to the data) and val (the value of the data with appropriate type). The NOT NULL constraint is imposed on all the columns. The xmldoc table stores the PDB ID's, docid's, the original PDBMLplus files, keywords (kwd) and the last modified time (mtime). Its docid column is referenced from the XPath-based tables as foreign keys.
| Column . | Type . | Example . |
|---|---|---|
| Container tables | `E:/datablock/entityCategory/entity' | |
| docid | INTEGER | 68 271 |
| pstart | INTEGER | 718 |
| pend | INTEGER | 737 |
| Content tables | `/datablock/entityCategory/entity/pdbx_ec' | |
| docid | INTEGER | 68 271 |
| pos | INTEGER | 727 |
| val | typeOfData | '1.1.3.9' |
| Xmldoc | xmldoc (this is the only instance) | |
| pdbid | TEXT | '1gof' |
| docid | INTEGER | 68 271 |
| doc | XML | '<datablock> … </datablock>' |
| kwd | TEXT | 'oxidation reduction … ' |
| mtime | TIMESTAMP | 2009-11-29 00:11:45+09 |
| Column . | Type . | Example . |
|---|---|---|
| Container tables | `E:/datablock/entityCategory/entity' | |
| docid | INTEGER | 68 271 |
| pstart | INTEGER | 718 |
| pend | INTEGER | 737 |
| Content tables | `/datablock/entityCategory/entity/pdbx_ec' | |
| docid | INTEGER | 68 271 |
| pos | INTEGER | 727 |
| val | typeOfData | '1.1.3.9' |
| Xmldoc | xmldoc (this is the only instance) | |
| pdbid | TEXT | '1gof' |
| docid | INTEGER | 68 271 |
| doc | XML | '<datablock> … </datablock>' |
| kwd | TEXT | 'oxidation reduction … ' |
| mtime | TIMESTAMP | 2009-11-29 00:11:45+09 |
There are two kinds of XPath-based tables for storing PDBMLplus data. The container tables contain the docid (PDB ID consisting of four alphanumerical characters interpreted as a radix 36 number), pstart (the pointer to the element) and pend (the maximum value of the pointers under that element). The table names of the container tables are XPaths prefixed with `E:'. The content tables contain docid, pos (the pointer to the data) and val (the value of the data with appropriate type). The NOT NULL constraint is imposed on all the columns. The xmldoc table stores the PDB ID's, docid's, the original PDBMLplus files, keywords (kwd) and the last modified time (mtime). Its docid column is referenced from the XPath-based tables as foreign keys.
The container table corresponding to an XML element contains three columns indicating the entry and the region of the element. Here, the region is defined as a pair of pointers, one to that element and the other to the last element under that element in the XML hierarchy of the document. In the example of Figure 2, the element for `/datablock/entryCategory/entry' of the PDB entry 1A00 would be represented as a triple (59616, 3, 4) where `59616' is the PDB ID 1A00 interpreted as a number of radix 36, and `3' is the pointer to the element, and `4' is the maximum value of the pointers under this element (in this case, this is the pointer to the attribute `/datablock/entryCategory/entry/@id'). This triple is stored in the table named `E:/datablock/entryCategory/entry'. The table names of container tables are always prefixed with `E:' as can be seen in this example. In a similar manner, in the table named `E:/datablock/struct_asymCategory/struct_asym,' triples such as (59616,6,9) and (59616, 10, 13) are present.
The content table corresponding to an attribute or a PCData contains three columns indicating the entry, the pointer to the data, and the data itself (i.e. attribute value or PCData). In the example of Figure 2, the value `1A00' for the XPath `/datablock/entryCategory/entry/@id' would be represented as a triple (59616, 4, `1A00') and is stored in the table named `/datablock/entryCategory/entry/@id'. Note that the `E:' prefix is absent from the names of content tables.
When an element contains a PCData directly under it, there are two tables with the same table names except for the `E:' prefix. For example, in Figure 2, the XML element `/datablock/struct_asymCategory/struct_asym/entity_id' containing the PCData `1' corresponds to the container table named `E:/datablock/struct_asymCategory/struct_asym/entity_id' containing a triple (docid, pstart, pend) = (59616, 8, 9) and to the content table named `/datablock/struct_asymCategory/struct_asym/entity_id' containing a triple (docid, pos, val) = (59616, 9, `1'). However, if an element can only contain a PCData without any attributes or other elements under it, the corresponding container table is redundant in the sense that it is not needed for retrieving meaningful information. Therefore, to save the required disk space, we do not keep such container tables. Whether or not an element can contain only a PCData can be determined from the PDBMLplus schema. In the above example, the container table `E:/datablock/struct_asymCategory/struct_asym/entity_id' does not actually exist in the working version of PDBj Mine.
Table definitions based on XML schema
As expected, there are a large number of tables required for storing PDBMLplus data. Nevertheless, owing to the simplicity of PDBMLplus schema and the basic structures of the relational tables, it is possible to generate the table definitions automatically from the PDBMLplus schema. Currently, there are 5368 XPath-based tables in PDBj Mine. The definitions for the container tables are identical for all elements except for the table names. The definitions for the content tables may be different for different tables due to the type of the data. Nevertheless, the type information in the PDBMLplus schema can be easily converted and transferred to relational table definitions. Since table names can be rather long, we also make a short-hand alias for each table as a view. For example, the table `E:/datablock/entryCategory/entry' can be referred to as `E://entry', and the table `/datablock/entryCategory/entry/@id' as `//entry/@id', etc. Unambiguous assignment of these short-hand notations is possible due to the regular structure of the PDBMLplus schema.
Terms for keyword search
The basic data types for content tables include integer, numeric (real number), date and text. For keyword search, however, we need to manually specify appropriate tables suitable for such a purpose. In the current implementation, we specify 23 PDBMLplus categories (out of 333) for keyword search. That is, all tables in these categories are indexed for keyword search if their data type is text. One exception is the identifier of the PubMed abstract database (http://www.ncbi.nlm.nih.gov/PubMed/), which is originally defined as integer but indexed as keywords (PubMed abstracts themselves are not available in PDBML or PDBMLplus).
Auxiliary tables for frequent queries and updates
In addition to the tables described so far, we also compile a table that summarizes the basic information which is queried frequently. This table, named `brief_summary', contains all the data needed for keyword search and Advanced Search (described below).
To facilitate managing the weekly update of PDBMLplus files, we also define a table named `xmldoc' that contains PDB ID, docid and, modified date of PDBMLplus files (Table 2). Thus, each row of the xmldoc table corresponds to one PDB entry, and the docid column of this table is referenced from all XPath-based tables using foreign keys. When a row in the xmldoc table is deleted, all data of the corresponding entry in other tables are automatically deleted. Therefore, when a PDBMLplus file is updated, we first delete the corresponding entry in the xmldoc table only, and then reload the updated data into the RDB. Although this strategy for updating entries is rather primitive, this is preferred to avoid complications regarding the manipulation of pointers for XPaths in many tables.
For convenience, we also included in the xmldoc table the raw PDBMLplus contents so that a user can retrieve the data using XPath expressions. However, in the current implementation, complex queries directly using XPath are not efficient (A few sample queries using XPath expressions can be found at http://doc.pdbj.org/help?PDBj%20Mine). Thus, this functionality should be used only for simple retrieval of PDBMLplus components.
Implementation
Relational database
As the back-end database management system (DBMS), we use PostgreSQL 8.4 (http://www.postgresql.org/), which has been compiled with XML-related functionality enabled. At the time of this writing, the version 8.4.4 is the latest one in the version 8.4 series of PostgreSQL (http://www.postgresql.org/ftp/source/v8.4.4/). We made minor modifications in the source code of PostgreSQL so that it can handle long table names.
The programs for converting PDBMLplus Schema to relational table definitions and for decomposing and loading PDBMLplus documents are written in the OCaml language (http://caml.inria.fr/) with supporting libraries to manipulate XML files and to interact with PostgreSQL. Several shell scripts are also employed to manage the weekly updates of PDB data.
Web interface
The user interface for PDBj Mine through the WWW is available at PDBj's web site (Figure 3). This web interface is implemented using Java servlets which accept input queries, and hand them to the back-end RDB and then convert the results into a simple XML format which is further processed with XML stylesheets to yield the final HTML pages. Depending on the type of queries, the workflow branches into six categories: (i) `Quick' searches using either PDB ID or keywords; (ii) advanced search with various conditions to filter the entries; (iii) SQL queries where the user can type in raw SQL expressions; (iv) author search to find entries deposited by particular authors; (v) subsequence search where the user can specify a short segment of a polymer sequence contained in PDB entries; and (vi) Search for most recently updated or newly added entries.
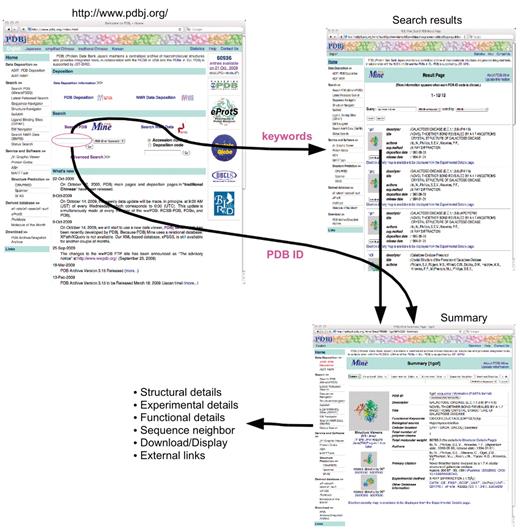
Entry retrieval and keyword search. From the PDBj top page, the user can input PDB ID, keywords, author names and subsequence of polymers (top left). When there are multiple hits to the query, a list of matching entries (search results) is returned (top right). When only one hit is found as in the case for PDB ID search or when an entry is selected in the search results, the summary page for the corresponding entry is shown (bottom right) from which other detailed pages (Structural, Experimental and Functional Details as well as Download/Display, and External Links) are linked.
As a result of a query, a list of PDB entries is usually obtained. The user can narrow down the result by adding PDB ID's, keywords, author names or subsequences. To facilitate viewing large result lists, the user can sort the list by PDB ID, deposition/release date or keyword relevance score. The keyword relevance score is calculated as the number of matching terms divided by the logarithm of the number of unique words in the keywords list of an entry (see the PostgreSQL manual).
For each PDB entry, there are dedicated summary and details pages (Figure 3). These pages, except for the `sequence neighbor' page, are generated from the original PDBMLplus files which are processed with XML stylesheets. The sequence neighbor page is dynamically generated from the result of a BLAST (7) search (This process usually takes less than a few seconds).
Web services
The RESTful web services are also provided. There are two services. One is a retrieval of PDBMLplus component of each entry based on an XPath expression embedded in a URL (access http://service.pdbj.org/mine/xpath for a brief instruction).
The other is the SQL search that accepts any valid SQL queries and returns the results in one of XML, CSV (comma-separated values), TSV (tab-separated values) or plain text formats (access http://service.pdbj.org/mine/sql for a brief instruction).
Database dumps
The database dump files are provided at our FTP server (ftp://ftp.pdbj.org/mine). The dump of the whole database is updated every week. The differential data for each week is also provided. Thus, users can construct a mirror of PDBj Mine provided that they have a customized PostgreSQL installed. For the compatibility, the version 8.4 series of PostgreSQL must be used (see the Relational database section) at the time of this writing. More detailed instructions are provided at the above FTP site.
Database statistics
As of 21July 2010, there were 66 633 entries in the PDB. The plain PDBMLplus files (without atomic coordinates) amounted to ∼55 GB. The size of the PDBj Mine database, including auxiliary tables and indexes, was ∼112 GB in total. Out of 5368 XPath-based tables, the most populated tables contained 40 331 637 rows and 4.4 GB of data (these were items of the pdbx_poly_seq_scheme category). On average, there were 213 511 rows per table but the median was 0 (corresponding to empty tables). Considering the heterogeneity and historical development of the PDB data, this is not surprising (8).
Examples
Entry retrieval and keyword search
Simple entry retrieval and keyword search can be performed from PDBj's top page (http://www.pdbj.org/). In addition, a short segment of polymer sequences and author names can be specified in the same input form. When a PDB ID is given in the form (Figure 3), a summary of the entry is returned, from which pages for structural details, experimental details functional details, sequence neighbors, download/display and external links are linked. In addition, interactive molecular graphics are also provided through either jV (9) or Jmol (http://www.jmol.org/). When keywords are input, a list of entries are displayed in a result page from which the user is directed to the summary page of each entry in the list.
Advanced Search
The Advanced Search interface available at http://service.pdbj.org/mine/advanced.html provides a range of conditions the user can specify to find desired entries. The conditions provided include release and deposition dates, citation information such as author names, journal titles and polymer types, ligands, cross-referenced database entries, experimental techniques, etc. This interface is implemented with a materialized view in the back-end database (i.e. the `brief_summary' table) to support quick response to queries. We expect that casual users can satisfy most of their needs by using this interface. Nevertheless, as our experience with user interaction increases, the Advanced Search interface will be improved accordingly.
SQL search
When users cannot satisfy their needs by using the interfaces mentioned above, they can resort to the SQL search interface available at http://service.pdbj.org/mine/ (entered by clicking the `PDBj Mine' logo on the PDBj top page). Doing so requires some understanding of the underlying structure of PDBMLplus and the RDB of PDBj Mine. Due to the relatively simple structure of the PDBMLplus schema, SQL queries can be constructed in a systematic manner. Here, we provide two examples based on the `entityCategory' element of PDBMLplus files (Figure 4A). Under this element, there are `entity' elements each of which describes a molecular entity contained in the PDBMLplus file. The following examples are rather low-level usage of the database, but the user can have the full control of the query. The equivalent queries can be greatly simplified by using the `category' views described in Appendix 1.

SQL query examples. (A) An example of a PDBMLplus document. (B) An example SQL query that retrieves the PDB ID and the description of molecular entities with EC number `1.1.1.1' annotated in the PDBMLplus file. (C) An example SQL query that retrieves the PDB ID and total molecular weight of polymers. Other examples of SQL queries are provided at http://doc.pdbj.org/help?PDBj%20Mine%3aSQL%20Queries.
To retrieve a list of PDB ID's and the descriptive names of entries with the EC (enzyme commission) number 1.1.1.1, the SQL query as in Figure 4B can be performed. First, PDB ID is selected from the `brief_summary' table to which three other tables are joined (the xmldoc table can be also used in place of brief_summary). The first joined table is `E://entity' which is a short-hand for `E:/datablock/entityCategory/entity' as mentioned above. The join condition is the equivalence of the `docid' column in brief_ summary and `E://entity' tables. To filter by the EC number, we join the table `//entity/pdbx_ec' which contains the desired information. This element is under the `entity' element, so that the join condition is the equivalence of `docid' as well as the pointer (`pos') column of the former being in the region of the latter (specified as `p1.pos BETWEEN e.pstart AND e.pend' in Figure 4B). To return the descriptive name of the entry, we also join the `//entity/pdbx_description' table in the same manner as the `//entity/pdbx_ec' table. Since we want the description of the entity with the specified EC number, the elements `pdbx_ec' and `pdbx_description' must be under the same `entity' element. This condition is naturally satisfied by requiring that the pointers to these elements be in the same region (spanned by `e.pstart' and `e.pend'). Finally, only those with EC number 1.1.1.1 are filtered by the WHERE clause (Figure 4B).
The next example is for retrieving a list of PDB ID's and the total molecular weights of polymers in the decreasing order of the latter (Figure 4C). In this query, five tables are joined together. First, in the same way as the previous example, the brief_summary and `E://entity' tables are joined. Next, three tables `//entity/type' (for filtering whether the entity is a polymer or not), `//entity/pdbx_number_of_molecules' (for counting the number of the entity in the entry), and `//entity/formula_weight' (for the molecular weight of the single entity) are joined with the same condition as in the previous query, that is, all being under the same `entity' element. Then, only those with `polymer' type are filtered in the WHERE clause, and finally, the results are aggregated in terms of PDB ID in the GROUP BY clause. The returned components are the PDB ID's and the total molecular weights computed with the standard SQL aggregate function SUM.
A number of simple as well as complex examples of SQL queries are provided at the PDBj help page (http://doc.pdbj.org/help). These examples may serve as templates for user's specific queries, and also as a tutorial for PDBj Mine.
Benchmark
We provide benchmark results comparing PDBj Mine with xPSSS, the previous web interface for PDBj based on a native XML DBMS. For most of the basic queries such as PDB ID pattern search, keyword search and various combinations in Advanced Search, the execution speed of PDBj Mine is comparable with that of xPSSS (Figure 5A). In 65 out of 94 equivalent queries, PDBj Mine performed better than xPSSS. Even for some relatively complicated queries in Advanced Search where xPSSS performs poorly, the performance of PDBj Mine is rather stable and robust.
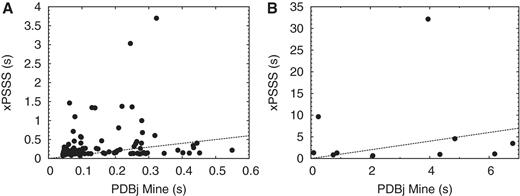
Benchmark of PDBj Mine compared with xPSSS. (A) Comparison of execution time for basic queries involving PDB ID search, keyword search and Advanced Search. (B) Comparison of execution time for SQL search in PDBj Mine and XQuery search in xPSSS. The numbers are the execution time (in seconds) for equivalent queries in PDBj Mine (abscissa) and xPSSS (ordinate). The xPSSS interface is based on a commercial native XML DBMS whereas PDBj Mine is based on a free open-source relational DBMS (PostgreSQL). For the details of the benchmark queries, see http://doc.pdbj.org/help?PDBj%20Mine.
We also compared more complex queries using SQL for PDBj Mine and XQuery for xPSSS. For half of the 10 equivalent queries tested, PDBj Mine performed better than xPSSS (Figure 5B). For the query in which PDBj Mine performed most poorly, the execution time was 6.8 s which was two times that of xPSSS. On the other hand, one query that took 32 s with xPSSS finished in 4 s with PDBj Mine. We conclude that the performance of PDBj Mine is consistent and robust although it is not always faster than xPSSS. We note that any queries that can be executed by using XQuery can be also executed by using SQL and vice versa. Thus, the main advantage of PDBj Mine over xPSSS is the robust and consistent performance.
Discussion
We have described the design and implementation as well as some examples of PDBj Mine. Here, we discuss some issues underlying the choices of our strategy.
The canonical data format for the wwPDB is the mmCIF format (2), which is in turn based on the STAR syntax (10) (STAR is to mmCIF what XML is to PDBML). The mmCIF format is designed for both humans and machines to read and interpret the contents of PDB data as unambiguously as possible. Although mmCIF does achieve its goal, the format is inherently limited to the PDB data so that specialized softwares are necessary to manipulate mmCIF files. On the other hand, PDBML or PDBMLplus files, being XML formats, can be handled with a wide range of softwares and are easily extensible (although PDBML files somewhat sacrifice the ease of reading by humans). Indeed, it is these features of PDBML (or XML) that motivated us to extend the PDBML format by adding our own annotations as well as annotations extracted from other databases. Thus, the use of PDBMLplus is beneficial to both the users and the developers.
Since we construct a database based on XML-formatted PDBMLplus files, it is natural to employ a native XML DBMS. We have provided the xPSSS interface based on a commercial XML DBMS before we have developed PDBj Mine which is based on a relational DBMS. The xPSSS interface served well for most cases. Nevertheless, we have been confronted with a number of problems. First, based on a commercial DBMS, it was not practical to construct mirror sites of PDBj by simply copying the softwares and data due to the license issues. This would not be a problem if we had used an open-source, free implementation of XML DBMS. However, as far as we have tested, there were no XML DBMSes that could meet our demand (mainly the amount of data and performance of queries). It appears that the technology for native XML databases is not yet mature at present so that the performance of some complex queries is not satisfactory (this applies even to commercial products to some extent). In contrast, RDBs are so mature and highly stable that non-commercial implementations can serve well in a demanding environment. For common queries, the current implementation of PDBj Mine exhibits a performance comparative to xPSSS, and for some complex queries, the performance of PDBj Mine far exceeds that of xPSSS. Thus, while the XML-based PDBMLplus files are convenient for adding annotations, RDB is more suitable for providing search services.
PDBML is derived from mmCIF which has been designed to be suitable for RDBs. In fact, each mmCIF category may be defined as a table in a RDB. However, we did not employ such design since it may become necessary in the future to define more deeply nested XML elements and multiple items with the same names under the same elements to integrate PDBML with other database resources. With the XML-oriented design of PDBj Mine, it is a trivial matter to add any XML-based data into PDBMLplus and expand PDBj Mine without altering the basic design. Nevertheless, for convenience, we also provide mmCIF category-like tables as views (see Appendix 1). We believe that it is a good compromise between the flexibility of XML and the robustness of relational DBMSs.
Funding
Grant-in-aid from the Institute for Bioinformatics Research and Development, Japan Science and Technology Agency (JST). Funding for open access charge: the Institute for Bioinformatics Research and Development, Japan Science and Technology Agency.
Conflict of interset. None declared.
References
Appendix 1
Category views
The basic database structure of PDBj Mine is based on PDBMLplus and each XML data corresponding to an mmCIF category item is saved in a distinct table. Although this design has an advantage in that it preserves the original structure of PDBMLplus files, it can easily be tedious to write SQL programs that combine many items. To circumvent this problem, we defined a view for each mmCIF category (referred to as `category view' in the following) which combines all items in that category. A schematic definition of a category view is given in Figure 6A. Sample SQL queries corresponding to those of Figures 4B and C are provided in Figures 6B and C, respectively.

Category views. (A) Schematic definition of a category view. (B) An SQL query equivalent to that of Figure 4B. (C) An SQL query equivalent to that of Figure 4C.
Although category views are easier to use compared with the XPath-based tables, a few caveats should be reminded. First, many columns of the category views are empty (i.e. containing NULL values) so that some queries might be polluted by meaningless values. Second, some views combine many items and/or merges very big tables so that certain queries can be significantly slow. If only a few columns of a category view are required, then the use of the XPath-based tables is recommended.



